Expression of Proteolytic Enzymes by Small Cell Lung Cancer Circulating Tumor Cell Lines
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Is Circulating HER-2 More Than Just a Tumor Marker?
Vol. 7, 2605–2607, September 2001 Clinical Cancer Research 2605 Editorial Is Circulating HER-2 More Than Just a Tumor Marker? Jose Baselga1 chemotherapy; and the selection patients for trastuzumab ther- Medical Oncology Service, Vall d’Hebron University Hospital, apy and the monitoring of response to trastuzumab (8, 9). Barcelona 08035, Spain. In this regard what does the study by Hayes et al. (10) add to our current knowledge? In this study, 242 patients who were enrolled in Cancer and Leukemia Group B (CALGB) prospec- HER2/neu (also known as neu and as c-erbB-2) is a proto- tive therapeutic trials for metastatic breast cancer were assayed oncogene of the EGF2 receptor family of receptor tyrosine for circulating ECD/HER-2 using a commercially available kinases (1). HER2/neu encodes a M 185,000 transmembrane r sandwich enzyme immunoassay (10). In their study, elevated glycoprotein receptor (HER2, or c-erbB-2) that has partial ho- ECD/HER-2 levels were observed in 37% of patients and were mology with the other members of the EGF receptor family, and associated with a shorter overall survival. However, in a multi- which also includes the EGF receptor (also called HER1), variate analysis, ECD/HER-2 did not independently correlate HER3, and HER4. These receptors are composed of an extra- with survival. Furthermore, ECD/HER-2 levels were not pre- cellular binding domain, a transmembrane lipophilic segment, dictive for time to progression and for response to megestrol and an intracellular protein tyrosine kinase domain with a reg- acetate or chemotherapy, including a subgroup of patients ulatory carboxyl terminal segment (2). -

Supporting Online Material
1 2 3 4 5 6 7 Supplementary Information for 8 9 Fractalkine-induced microglial vasoregulation occurs within the retina and is altered early in diabetic 10 retinopathy 11 12 *Samuel A. Mills, *Andrew I. Jobling, *Michael A. Dixon, Bang V. Bui, Kirstan A. Vessey, Joanna A. Phipps, 13 Ursula Greferath, Gene Venables, Vickie H.Y. Wong, Connie H.Y. Wong, Zheng He, Flora Hui, James C. 14 Young, Josh Tonc, Elena Ivanova, Botir T. Sagdullaev, Erica L. Fletcher 15 * Joint first authors 16 17 Corresponding author: 18 Prof. Erica L. Fletcher. Department of Anatomy & Neuroscience. The University of Melbourne, Grattan St, 19 Parkville 3010, Victoria, Australia. 20 Email: [email protected] ; Tel: +61-3-8344-3218; Fax: +61-3-9347-5219 21 22 This PDF file includes: 23 24 Supplementary text 25 Figures S1 to S10 26 Tables S1 to S7 27 Legends for Movies S1 to S2 28 SI References 29 30 Other supplementary materials for this manuscript include the following: 31 32 Movies S1 to S2 33 34 35 36 1 1 Supplementary Information Text 2 Materials and Methods 3 Microglial process movement on retinal vessels 4 Dark agouti rats were anaesthetized, injected intraperitoneally with rhodamine B (Sigma-Aldrich) to label blood 5 vessels and retinal explants established as described in the main text. Retinal microglia were labelled with Iba-1 6 and imaging performed on an inverted confocal microscope (Leica SP5). Baseline images were taken for 10 7 minutes, followed by the addition of PBS (10 minutes) and then either fractalkine or fractalkine + candesartan 8 (10 minutes) using concentrations outlined in the main text. -
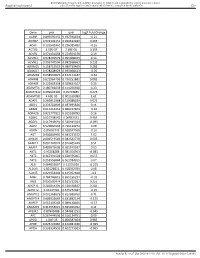
Gene Pval Qval Log2 Fold Change AAMP 0.895690332 0.952598834
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) Gut Gene pval qval Log2 Fold Change AAMP 0.895690332 0.952598834 -0.21 ABI3BP 0.002302151 0.020612283 0.465 ACHE 0.103542461 0.296385483 -0.16 ACTG2 2.99E-07 7.68E-05 3.195 ACVR1 0.071431098 0.224504378 0.19 ACVR1C 0.978209579 0.995008423 0.14 ACVRL1 0.006747504 0.042938663 0.235 ADAM15 0.158715519 0.380719469 0.285 ADAM17 0.978208929 0.995008423 -0.05 ADAM28 0.038932876 0.152174187 -0.62 ADAM8 0.622964796 0.790251882 0.085 ADAM9 0.122003358 0.329623107 0.25 ADAMTS1 0.180766659 0.414256926 0.23 ADAMTS12 0.009902195 0.05703885 0.425 ADAMTS8 4.60E-05 0.001169089 1.61 ADAP1 0.269811968 0.519388039 0.075 ADD1 0.233702809 0.487695826 0.11 ADM2 0.012213453 0.066227879 -0.36 ADRA2B 0.822777921 0.915518785 0.16 AEBP1 0.010738542 0.06035531 0.465 AGGF1 0.117946691 0.320915024 -0.095 AGR2 0.529860903 0.736120272 0.08 AGRN 0.85693743 0.928047568 -0.16 AGT 0.006849995 0.043233572 1.02 AHNAK 0.006519543 0.042542779 0.605 AKAP12 0.001747074 0.016405449 0.51 AKAP2 0.409929603 0.665919397 0.05 AKT1 0.95208288 0.985354963 -0.085 AKT2 0.367391504 0.620376005 0.055 AKT3 0.253556844 0.501934205 0.07 ALB 0.064833867 0.21195036 -0.315 ALDOA 0.83128831 0.918352939 0.08 ALOX5 0.029954404 0.125352668 -0.3 AMH 0.784746815 0.895196237 -0.03 ANG 0.050500474 0.181732067 0.255 ANGPT1 0.281853305 0.538528647 0.285 ANGPT2 0.43147281 0.675272487 -0.15 ANGPTL2 0.001368876 0.013688762 0.71 ANGPTL4 0.686032669 0.831882134 -0.175 ANPEP 0.019103243 0.089148466 -0.57 ANXA2P2 0.412553021 0.665966092 0.11 AP1M2 0.87843088 0.944681253 -0.045 APC 0.267444505 0.516134751 0.09 APOD 1.04E-05 0.000587404 0.985 APOE 0.023722987 0.104981036 -0.395 APOH 0.336334555 0.602273505 -0.065 Sundar R, et al. -

Cerebrospinal Fluid Ctdna and Metabolites Are Informative
www.nature.com/scientificreports OPEN Cerebrospinal fuid ctDNA and metabolites are informative biomarkers for the evaluation of CNS germ cell tumors Takeshi Takayasu1,2, Mauli Shah1, Antonio Dono3, Yuanqing Yan3, Roshan Borkar4, Nagireddy Putluri4, Jay‑Jiguang Zhu3,5, Seiji Hama2, Fumiyuki Yamasaki2*, Hidetoshi Tahara6, Kazuhiko Sugiyama7, Kaoru Kurisu2, Yoshua Esquenazi3,5,8 & Leomar Y. Ballester1,3,5* Serum and cerebrospinal fuid (CSF) levels of α‑fetoprotein and β‑subunit of human chorionic gonadotropin are used as biomarkers for the management of central nervous system (CNS) germ cell tumors (GCTs). However, additional discriminating biomarkers are required. Especially, biomarkers to diferentiate non‑germinomatous germ cell tumors (NGGCTs) from germinomas are critical, as these have a distinct prognosis. We investigated CSF samples from 12 patients with CNS‑GCT patients (8 germinomas and 4 NGGCTs). We analyzed circulating tumor DNA (ctDNA) in CSF to detect mutated genes. We also used liquid chromatography‑mass spectrometry to characterize metabolites in CSF. We detected KIT and/or NRAS mutation, known as frequently mutated genes in GCTs, in 3/12 (25%) patients. We also found signifcant diferences in the abundance of 15 metabolites between control and GCT, with unsupervised hierarchical clustering analysis. Metabolites related to the TCA cycle were increased in GCTs. Urea, ornithine, and short‑chain acylcarnitines were decreased in GCTs. Moreover, we also detected several metabolites (e.g., betaine, guanidine acetic acid, and 2‑aminoheptanoic acid) that displayed signifcant diferences in abundance in patients with germinomas and NGGCTs. Our results suggest that ctDNA and metabolites in CSF can serve as novel biomarkers for CNS‑GCTs and can be useful to diferentiate germinomas from NGGCTs. -

R&D Assay for Alzheimer's Disease
R&DR&D assayassay forfor Alzheimer’sAlzheimer’s diseasedisease Target screening⳼ Ⲽ㬔 antibody array, ᢜ⭉㬔 ⸽ἐⴐ Amyloid β-peptide Alzheimer’s disease⯸ ኸᷠ᧔ ᆹ⸽ inhibitor, antibody, ELISA kit Surwhrph#Surilohu#Dqwlerg|#Duud| 6OUSFBUFE 1."5SFBUFE )41 $3&# &3, &3, )41 $3&# &3, &3, 壤伡庰䋸TBNQMF ɅH 侴䋸嵄䍴䋸BOBMZUFT䋸䬱娴哜塵 1$ 1$ 1$ 1$ 5IFNPTUSFGFSFODFEBSSBZT 1$ 1$ QQ α 34, .4, 503 Q α 34, .4, 503 %SVHTDSFFOJOH0òUBSHFUFòFDUT0ATHWAY涭廐 6OUSFBUFE 堄币䋸4BNQMF侴䋸8FTUFSOPS&-*4"䍘䧽 1."5SFBUFE P 8FTUFSOCMPU廽喜儤应侴䋸0, Z 4VCTUSBUF -JHIU )31DPOKVHBUFE1BO "OUJQIPTQIPUZSPTJOF .FBO1JYFM%FOTJUZ Y $BQUVSF"OUJCPEZ 5BSHFU"OBMZUF "SSBZ.FNCSBOF $3&# &3, &3, )41 .4, Q α 34, 503 Human XL Cytokine Array kit (ARY022, 102 analytes) Adiponectin,Aggrecan,Angiogenin,Angiopoietin-1,Angiopoietin-2,BAFF,BDNF,Complement,Component C5/C5a,CD14,CD30,CD40L, Chitinase 3-like 1,Complement Factor D,C-Reactive Protein,Cripto-1,Cystatin C,Dkk-1,DPPIV,EGF,EMMPRIN,ENA-78,Endoglin, Fas L,FGF basic,FGF- 7,FGF-19,Flt-3 L,G-CSF,GDF-15,GM-CSF,GRO-α,Grow th Hormone,HGF,ICAM-1,IFN-γ,IGFBP-2,IGFBP-3, IL-1α,IL-1β, IL-1ra,IL-2,IL-3,IL-4,IL- 5,IL-6,IL-8, IL-10,IL-11,IL-12, IL-13,IL-15,IL-16,IL-17A,IL-18 BPa,IL-19,IL-22, IL-23,IL-24,IL-27, IL-31,IL-32α/β/γ,IL-33,IL-34,IP-10,I-TAC,Kallikrein 3,Leptin,LIF,Lipocalin-2,MCP-1,MCP-3,M-CSF,MIF,MIG,MIP-1α/MIP-1β,MIP-3α,MIP-3β,MMP-9, Myeloperoxidase,Osteopontin, p70, PDGF-AA, PDGF-AB/BB,Pentraxin-3, PF4, RAGE, RANTES,RBP4,Relaxin-2, Resistin,SDF-1α,Serpin E1, SHBG, ST2, TARC,TFF3,TfR,TGF- ,Thrombospondin-1,TNF-α, uPAR, VEGF, Vitamin D BP Human Protease (34 analytes) / -

(CEA) As a Serum Marker for Breast Cancer: a Prospective Longitudinal Study1
Vol. 7, 2357–2362, August 2001 Clinical Cancer Research 2357 A Re-Evaluation of Carcinoembryonic Antigen (CEA) as a Serum Marker for Breast Cancer: A Prospective Longitudinal Study1 Fiorella Guadagni,2 Patrizia Ferroni, Longitudinal monitoring of 53 metastatic patients undergo- Sandro Carlini, Sabrina Mariotti, Antonella Spila, ing chemotherapy demonstrated that, when positive, both Simona Aloe, Roberta D’Alessandro, CEA and CA 15.3 paralleled response to treatment, al- though CA 15.3 was a significantly more powerful marker Maria Daniela Carone, Americo Cicchetti, for determining response to treatment. The cost effective- Andrea Ricciotti, Irene Venturo, Pasquale Perri, ness ratio of CEA was clearly less favorable than that of Franco Di Filippo, Francesco Cognetti, CA 15.3. Claudio Botti, and Mario Roselli Conclusions: CEA monitoring should be considered an Laboratory of Clinical Pathology [F. G., S. M., A. S., S. A., R. D., expensive and inefficient method of follow-up evaluation for M. D. C.], III Department of Surgery [S. C., P. P.], I Department of breast cancer patients, and it provides no additional value Surgery [A. R., F. D. F., C. B.], and Department of Medical Oncology when used in combination with CA 15.3. [F. C., I. V.], Regina Elena Cancer Institute, Department of Experimental Medicine and Pathology, University of Rome “La Sapienza,” [P. F.]; Department of Hygiene and Public Health, INTRODUCTION Catholic University of Rome “Sacro Cuore,” [A. C.]; and Department CEA3 is one of the first tumor markers to be identified and of Surgery, University of Rome “Tor Vergata,” 00100 Rome, Italy [M. R.] characterized (1, 2). -

Table 6. Putative Genes Involved in the Utilization of Carbohydrates in G
Table 6. Putative genes involved in the utilization of carbohydrates in G. thermodenitrificans NG80-2 genome Carbohydrates* Enzymes Gene ID Glycerol Glycerol Kinase GT1216 Glycerol-3-phosphate dehydrogenase, aerobic GT2089 NAD(P)H-dependent glycerol-3-phosphate dehydrogenase GT2153 Enolase GT3003 2,3-bisphosphoglycerate-independentphosphoglycerate mutase GT3004 Triosephosphate isomerase GT3005 3-phosphoglycerate kinase GT3006 Glyceraldehyde-3-phosphate dehydrogenase GT3007 Phosphoglycerate mutase GT1326 Pyruvate kinase GT2663 L-Arabinose L-arabinose isomerase GT1795 L-ribulokinase GT1796 L-ribulose 5-phosphate 4-epimerase GT1797 D-Ribose Ribokinase GT3174 Transketolase GT1187 Ribose 5-phosphate epimerase GT3316 D-Xylose Xylose kinase GT1756 Xylose isomerase GT1757 D-Galactose Galactokinase GT2086 Galactose-1-phosphate uridyltransferase GT2084 UDP-glucose 4-epimerase GT2085 Carbohydrates* Enzymes Gene ID D-Fructose 1-phosphofructokinase GT1727 Fructose-1,6-bisphosphate aldolase GT1805 Fructose-1,6-bisphosphate aldolase type II GT3331 Triosephosphate isomerase GT3005 D-Mannose Mannnose-6 phospate isomelase GT3398 6-phospho-1-fructokinase GT2664 D-Mannitol Mannitol-1-phosphate dehydrogenase GT1844 N-Acetylglucosamine N-acetylglucosamine-6-phosphate deacetylase GT2205 N-acetylglucosamine-6-phosphate isomerase GT2204 D-Maltose Alpha-1,4-glucosidase GT0528, GT1643 Sucrose Sucrose phosphorylase GT3215 D-Trehalose Alpha-glucosidase GT1643 Glucose kinase GT2381 Inositol Myo-inositol catabolism protein iolC;5-dehydro-2- GT1807 deoxygluconokinase -

Supplementary Material DNA Methylation in Inflammatory Pathways Modifies the Association Between BMI and Adult-Onset Non- Atopic
Supplementary Material DNA Methylation in Inflammatory Pathways Modifies the Association between BMI and Adult-Onset Non- Atopic Asthma Ayoung Jeong 1,2, Medea Imboden 1,2, Akram Ghantous 3, Alexei Novoloaca 3, Anne-Elie Carsin 4,5,6, Manolis Kogevinas 4,5,6, Christian Schindler 1,2, Gianfranco Lovison 7, Zdenko Herceg 3, Cyrille Cuenin 3, Roel Vermeulen 8, Deborah Jarvis 9, André F. S. Amaral 9, Florian Kronenberg 10, Paolo Vineis 11,12 and Nicole Probst-Hensch 1,2,* 1 Swiss Tropical and Public Health Institute, 4051 Basel, Switzerland; [email protected] (A.J.); [email protected] (M.I.); [email protected] (C.S.) 2 Department of Public Health, University of Basel, 4001 Basel, Switzerland 3 International Agency for Research on Cancer, 69372 Lyon, France; [email protected] (A.G.); [email protected] (A.N.); [email protected] (Z.H.); [email protected] (C.C.) 4 ISGlobal, Barcelona Institute for Global Health, 08003 Barcelona, Spain; [email protected] (A.-E.C.); [email protected] (M.K.) 5 Universitat Pompeu Fabra (UPF), 08002 Barcelona, Spain 6 CIBER Epidemiología y Salud Pública (CIBERESP), 08005 Barcelona, Spain 7 Department of Economics, Business and Statistics, University of Palermo, 90128 Palermo, Italy; [email protected] 8 Environmental Epidemiology Division, Utrecht University, Institute for Risk Assessment Sciences, 3584CM Utrecht, Netherlands; [email protected] 9 Population Health and Occupational Disease, National Heart and Lung Institute, Imperial College, SW3 6LR London, UK; [email protected] (D.J.); [email protected] (A.F.S.A.) 10 Division of Genetic Epidemiology, Medical University of Innsbruck, 6020 Innsbruck, Austria; [email protected] 11 MRC-PHE Centre for Environment and Health, School of Public Health, Imperial College London, W2 1PG London, UK; [email protected] 12 Italian Institute for Genomic Medicine (IIGM), 10126 Turin, Italy * Correspondence: [email protected]; Tel.: +41-61-284-8378 Int. -

An Investigation on Catalysis of Acylaminoacyl Peptidases
An investigation on catalysis of acylaminoacyl peptidases PhD Thesis András László Kiss Doctorate School of Biology School leader: Prof. Anna Erdei Structural Biochemistry Programme Programme leader: Prof. László Gráf Supervisor: Prof. László Polgár Institute of Enzymology, Biological Research Center, Hungarian Academy of Sciences Budapest 2007 1. Introduction Serine peptidases contain two residues at the active site in addition to the catalytic triad (His, Asp, Ser), which form a cavity called “oxyanion hole” that accommodates the negatively charged oxyanion in the transition state of the catalysis and donate two H- bonds. Enzymes of the prolyl oligopeptidase (POP) family (acylaminoacyl peptidase, prolyl oligopeptidase, dipeptidyl peptidase IV and oligopeptidase B) are extensively studied. These enzymes are larger than classical serine peptidases and are composed of a peptidase domain with an α/β hydrolase fold and a β-propeller domain. POP and oligopeptidase B are monomeric enzymes and endopeptidases, while the exopeptidase acylaminoacyl peptidase and dipeptidyl peptidase IV are tetrameric and dimeric enzymes, respectively. Acylaminoacyl peptidase (AAP) cleaves acylated amino acids from the N-terminus of the N-acylated peptides that plays important role in many biological and disease processes. Human AAP is encoded by the DNF15S2 locus on the short arm of chromosome 3 at the region 21, which suffers deletions in small cell lung carcinomas, and renal carcinomas, resulting in deficiency in the expression of the enzyme. Acylaminoacyl peptidase is also supposed to be involved in the degradation of oxidatively damaged proteins in cells and can be associated with various diseases where damaged proteins aggregate. Its involvement in cataract formation and in the breakdown of immunogenic formylmethionyl-peptides in the digestive tract after bacterial attack was also suggested. -

High-Resolution Mass Spectrometry-Based Approaches for the Detection and Quantification of Peptidase Activity in Plasma
molecules Article High-Resolution Mass Spectrometry-Based Approaches for the Detection and Quantification of Peptidase Activity in Plasma Elisa Maffioli 1,2 , Zhenze Jiang 3, Simona Nonnis 1,2 , Armando Negri 1,2, Valentina Romeo 1, Christopher B. Lietz 3, Vivian Hook 3,4, Giuseppe Ristagno 5, Giuseppe Baselli 6, Erik B. Kistler 7,8 , Federico Aletti 9, Anthony J. O’Donoghue 3,* and Gabriella Tedeschi 1,2,* 1 Department of Veterinary Medicine, University of Milano, 20133 Milano, Italy; elisa.maffi[email protected] (E.M.); [email protected] (S.N.); [email protected] (A.N.); [email protected] (V.R.) 2 Centre for Nanostructured Materials and Interfaces (CIMAINA), University of Milano, 20133 Milano, Italy 3 Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California San Diego, La Jolla, CA 92093, USA; [email protected] (Z.J.); [email protected] (C.B.L.); [email protected] (V.H.) 4 Department of Neurosciences, School of Medicine, University of California San Diego, La Jolla, CA 92093, USA 5 Department of Pathophysiology and Transplantation, University of Milan, 20133 Milan, Italy; [email protected] 6 Dipartimento di Elettronica, Informazione e Bioingegneria, Politecnico di Milano, 20133 Milan, Italy; [email protected] 7 Department of Anesthesiology & Critical Care, University of California San Diego, La Jolla, CA 92093, USA; [email protected] 8 Department of Anesthesiology & Critical Care, VA San Diego HealthCare System, San Diego, CA 92161, USA 9 Department of Bioengineering, University of California San Diego, La Jolla, CA 92093, USA; [email protected] * Correspondence: [email protected] (A.J.O.); [email protected] (G.T.); Tel.: +1-8585345360 (A.J.O.); +39-02-50318127 (G.T.) Academic Editor: Paolo Iadarola Received: 28 July 2020; Accepted: 4 September 2020; Published: 6 September 2020 Abstract: Proteomic technologies have identified 234 peptidases in plasma but little quantitative information about the proteolytic activity has been uncovered. -

A Preliminary Evaluation of Calcitonin and PDN-21 As Tumor Markers for Lung Cancer
Henry Ford Hospital Medical Journal Volume 37 Number 3 Article 31 9-1989 A Preliminary Evaluation of Calcitonin and PDN-21 as Tumor Markers for Lung Cancer J. J. Body J. C. Dumon J. P. Sculier G. Dabouis H. Lacroix See next page for additional authors Follow this and additional works at: https://scholarlycommons.henryford.com/hfhmedjournal Part of the Life Sciences Commons, Medical Specialties Commons, and the Public Health Commons Recommended Citation Body, J. J.; Dumon, J. C.; Sculier, J. P.; Dabouis, G.; Lacroix, H.; Libert, P.; Richez, M.; Bureau, G.; Mommen, P.; Raymakers, N.; Paesmans, M.; and Klastersky, J. (1989) "A Preliminary Evaluation of Calcitonin and PDN-21 as Tumor Markers for Lung Cancer," Henry Ford Hospital Medical Journal : Vol. 37 : No. 3 , 190-193. Available at: https://scholarlycommons.henryford.com/hfhmedjournal/vol37/iss3/31 This Article is brought to you for free and open access by Henry Ford Health System Scholarly Commons. It has been accepted for inclusion in Henry Ford Hospital Medical Journal by an authorized editor of Henry Ford Health System Scholarly Commons. A Preliminary Evaluation of Calcitonin and PDN-21 as Tumor Markers for Lung Cancer Authors J. J. Body, J. C. Dumon, J. P. Sculier, G. Dabouis, H. Lacroix, P. Libert, M. Richez, G. Bureau, P. Mommen, N. Raymakers, M. Paesmans, and J. Klastersky This article is available in Henry Ford Hospital Medical Journal: https://scholarlycommons.henryford.com/ hfhmedjournal/vol37/iss3/31 A Preliminary Evaluation of Calcitonin and PDN-21 as TUmor Markers for Lung Cancer J.J. Body,* J.C. Dumon,* J.P. -

HER2 Testing Fact Sheet
FACT SHEET Media: Krysta Pellegrino (650) 467-6800 Investor: Diane Schrick (650) 225-1599 Advocacy: Sonali Padhi (650) 467-0842 HER2 Testing Fact Sheet Testing tumors to determine their genetic makeup provides vital information about cancer at the cellular level. Together with physical characteristics, such as size, type and stage of the tumor, tumor marker testing can help determine the appropriate medicines needed to treat the disease most effectively. A variety of tests to identify breast and stomach (gastric) cancer tumor markers can be performed. Herceptin® (trastuzumab) is approved in combination with the chemotherapy drugs cisplatin, and either capecitabine or 5-fluorouracil, for metastatic HER2-positive stomach cancer or cancer of the gastroesophageal junction, in men and women who have not received prior medicines for their metastatic disease. Who Should Be Tested Guidelines from various organizations recommend people with breast cancer receive tumor marker testing, including the American Society of Clinical Oncology (ASCO), the College of American Pathologists (CAP) and the National Comprehensive Cancer Network (NCCN).1,2,3 No guidelines exist yet for stomach cancer testing. A person who had a biopsy (a tissue sample taken from the tumor for testing), but has not received a tumor marker test, may request it from his or her doctor. If a person is told their sample is no longer being stored, a new biopsy may be requested if the tumor is still present or if the cancer returns. People should ask their doctor for additional information about testing. HER2 Testing For Breast and Stomach Cancer Standard human epidermal growth factor receptor 2 (HER2) tests measure if there is a higher than normal number of HER2 genes present in tumor cells or if there is a higher than normal number of HER2 receptors on the surface of tumor cells.