Abbreviations: ABPC, Ampicillin
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Cusumano-Et-Al-2017.Pdf
International Journal of Infectious Diseases 63 (2017) 1–6 Contents lists available at ScienceDirect International Journal of Infectious Diseases journal homepage: www.elsevier.com/locate/ijid Rapidly growing Mycobacterium infections after cosmetic surgery in medical tourists: the Bronx experience and a review of the literature a a b c b Lucas R. Cusumano , Vivy Tran , Aileen Tlamsa , Philip Chung , Robert Grossberg , b b, Gregory Weston , Uzma N. Sarwar * a Albert Einstein College of Medicine, Montefiore Medical Center, Bronx, New York, USA b Division of Infectious Diseases, Department of Medicine, Albert Einstein College of Medicine, Montefiore Medical Center, Bronx, New York, USA c Department of Pharmacy, Nebraska Medicine, Omaha, Nebraska, USA A R T I C L E I N F O A B S T R A C T Article history: Background: Medical tourism is increasingly popular for elective cosmetic surgical procedures. However, Received 10 May 2017 medical tourism has been accompanied by reports of post-surgical infections due to rapidly growing Received in revised form 22 July 2017 mycobacteria (RGM). The authors’ experience working with patients with RGM infections who have Accepted 26 July 2017 returned to the USA after traveling abroad for cosmetic surgical procedures is described here. Corresponding Editor: Eskild Petersen, Methods: Patients who developed RGM infections after undergoing cosmetic surgeries abroad and who ?Aarhus, Denmark presented at the Montefiore Medical Center (Bronx, New York, USA) between August 2015 and June 2016 were identified. A review of patient medical records was performed. Keywords: Results: Four patients who presented with culture-proven RGM infections at the sites of recent cosmetic Mycobacterium abscessus complex procedures were identified. -

The Activity of Mecillinam Vs Enterobacteriaceae Resistant to 3Rd Generation Cephalosporins in Bristol, UK
The activity of mecillinam vs Enterobacteriaceae resistant to 3rd generation cephalosporins in Bristol, UK Welsh Antimicrobial Study Group Grŵp Astudio Wrthfiotegau Cymru G Weston1, KE Bowker1, A Noel1, AP MacGowan1, M Wootton2, TR Walsh2, RA Howe2 (1) BCARE, North Bristol NHS Trust, Bristol BS10 5NB (2) Welsh Antimicrobial Study Group, NPHS Wales, University Hospital of Wales, CF14 4XW Introduction Results Results Results Figure 1: Population Distributions of Mecillinam MICs for E. Figure 2: Population Distributions of MICs for ESBL- Resistance in coliforms to 3rd generation 127 isolates were identified by screening of which 123 were confirmed as resistant to CTX or coli (n=72), NON-E. coli Enterobacteriaceae (n=47) and multi- producing E. coli (n=67) against mecillinam or mecillinam + cephalosporins (3GC) is an increasing problem resistant strains (n=39) clavulanate CAZ by BSAC criteria. The majority of 3GC- both in hospitals and the community. Oral options MR Non-E. coli E. coli Mecalone Mec+Clav resistant strains were E. coli 60.2%, followed by for the treatment of these organisms is often 35 50 Enterobacter spp. 16.2%, Klebsiella spp. 12.2%, limited due to resistance to multiple antimicrobial 45 and others (Citrobacter spp., Morganella spp., 30 classes. Mecillinam, an amidinopenicillin that is 40 Pantoea spp., Serratia spp.) 11.4%. 25 available in Europe as the oral pro-drug 35 All isolates were susceptible to meropenem with s 20 30 e s pivmecillinam, is stable to many beta-lactamases. t e a t l a mecillinam the next most active agent with more l o 25 o s We aimed to establish the activity of mecillinam i s i 15 % than 95% of isolates susceptible (table). -
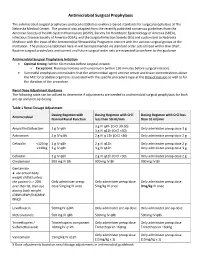
Antimicrobial Surgical Prophylaxis
Antimicrobial Surgical Prophylaxis The antimicrobial surgical prophylaxis protocol establishes evidence-based standards for surgical prophylaxis at The Nebraska Medical Center. The protocol was adapted from the recently published consensus guidelines from the American Society of Health-System Pharmacists (ASHP), Society for Healthcare Epidemiology of America (SHEA), Infectious Disease Society of America (IDSA), and the Surgical Infection Society (SIS) and customized to Nebraska Medicine with the input of the Antimicrobial Stewardship Program in concert with the various surgical groups at the institution. The protocol established here-in will be implemented via standard order sets utilized within One Chart. Routine surgical prophylaxis and current and future surgical order sets are expected to conform to this guidance. Antimicrobial Surgical Prophylaxis Initiation Optimal timing: Within 60 minutes before surgical incision o Exceptions: Fluoroquinolones and vancomycin (within 120 minutes before surgical incision) Successful prophylaxis necessitates that the antimicrobial agent achieve serum and tissue concentrations above the MIC for probable organisms associated with the specific procedure type at the time of incision as well as for the duration of the procedure. Renal Dose Adjustment Guidance The following table can be utilized to determine if adjustments are needed to antimicrobial surgical prophylaxis for both pre-op and post-op dosing. Table 1 Renal Dosage Adjustment Dosing Regimen with Dosing Regimen with CrCl Dosing Regimen with -

In Vitro Susceptibilities of Escherichia Coli and Klebsiella Spp. To
Jpn. J. Infect. Dis., 60, 227-229, 2007 Short Communication In Vitro Susceptibilities of Escherichia coli and Klebsiella Spp. to Ampicillin-Sulbactam and Amoxicillin-Clavulanic Acid Birgul Kacmaz* and Nedim Sultan1 Department of Central Microbiology and 1Department of Microbiology, Faculty of Medicine, Gazi University, Ankara, Turkey (Received January 30, 2007. Accepted April 13, 2007) SUMMARY: Ampicillin-sulbactam (A/S) and amoxicillin-clavulanic acid (AUG) are thought to be equally efficacious clinically against the Enterobacteriaceae family. In this study, the in vitro activities of the A/S and AUG were evaluated and compared against Escherichia coli and Klebsiella spp. Antimicrobial susceptibility tests were performed by standard agar dilution and disc diffusion techniques according to the Clinical and Laboratory Standards Institute (CLSI). During the study period, 973 strains were isolated. Of the 973 bacteria isolated, 823 were E. coli and 150 Klebsiella spp. More organisms were found to be susceptible to AUG than A/S, regardless of the susceptibility testing methodology. The agar dilution results of the isolates that were found to be sensitive or resistant were also compatible with the disc diffusion results. However, some differences were seen in the agar dilution results of some isolates that were found to be intermediately resistant with disc diffusion. In E. coli isolates, 17 of the 76 AUG intermediately resistant isolates (by disc diffusion), and 17 of the 63 A/S intermediately resistant isolates (by disc diffusion) showed different resistant patterns by agar dilution. When the CLSI breakpoint criteria are applied it should be considered that AUG and A/S sensitivity in E. coli and Klebsiella spp. -

Antimicrobial Stewardship Guidance
Antimicrobial Stewardship Guidance Federal Bureau of Prisons Clinical Practice Guidelines March 2013 Clinical guidelines are made available to the public for informational purposes only. The Federal Bureau of Prisons (BOP) does not warrant these guidelines for any other purpose, and assumes no responsibility for any injury or damage resulting from the reliance thereof. Proper medical practice necessitates that all cases are evaluated on an individual basis and that treatment decisions are patient-specific. Consult the BOP Clinical Practice Guidelines Web page to determine the date of the most recent update to this document: http://www.bop.gov/news/medresources.jsp Federal Bureau of Prisons Antimicrobial Stewardship Guidance Clinical Practice Guidelines March 2013 Table of Contents 1. Purpose ............................................................................................................................................. 3 2. Introduction ...................................................................................................................................... 3 3. Antimicrobial Stewardship in the BOP............................................................................................ 4 4. General Guidance for Diagnosis and Identifying Infection ............................................................. 5 Diagnosis of Specific Infections ........................................................................................................ 6 Upper Respiratory Infections (not otherwise specified) .............................................................................. -

Susceptibility of Pseudomonas Cepacia Isolated from Children with Cystic Fibrosis1
003 1-3998/86/2011-1174$02.00/0 PEDIATRIC RESEARCH Vol. 20, No. 1 1, 1986 Copyright O 1986 International Pediatric Research Foundation, Inc. Printed in (I.S.A. Decreased Baseline P-Lactamase Production and Inducibility associated with Increased Piperacillin Susceptibility of Pseudomonas cepacia Isolated from Children with Cystic Fibrosis1 CLAUDIO CHIESA,~PAULINE H. LABROZZI, AND STEPHEN C. ARONOFF Department of Pediatrics [C.C., P.H.L., S.C.A.], Case- Western Reserve University School ofMedicine and the Division of Pediatric Infectious Diseases [S.C.A.], Rainbow Babies and Children's Hospital, Cleveland, Ohio ABSTRACT. The incidence of pulmonary infections in creased since 1978 and is now recovered from 20% of patients children with cystic fibrosis caused by Pseudomonas ce- in some centers (3, 4). pacia, an organism which may possess an inducible 8- Sputum isolates of P. cepacia from children with cystic fibrosis lactamase, has increased since 1978. Seven of 13 sputum are resistant to most P-lactam agents in vitro. In an in vitro study isolates of P. cepacia from children with cystic fibrosis comparing the susceptibilities of 62 consecutive sputum isolates, were classified as inducible by quantitative enzyme produc- concentrations of 64 pg/ml of aztreonam and piperacillin were tion following preincubation with 100, 200, or 400 pg/ml required to inhibit 79 and 87.1 % of the bacterial population, of cefoxitin. The recovery of inducible strains tended to be respectively; 90% of the isolates were inhibited by 8 pg/ml of associated with recent ceftazidime therapy. Susceptibility ceftazidime (5). In vitro, ceftazidime has proven more active to aztreonam, ceftazidime, and piperacillin alone or com- against P. -

Mecillinam (FL 1060), a 6,3-Amidinopenicillanic Acid Derivative: Bactericidal Action and Synergy in Vitro L
ANTUCROBIAL AGENTS AND CHEzOTHrAPY, Sept. 1975, p. 271-276 Vol. 8, No. 3 Copyright 0 1975 American Society for Microbiology Printed in U.S.A. Mecillinam (FL 1060), a 6,3-Amidinopenicillanic Acid Derivative: Bactericidal Action and Synergy In Vitro L. TYBRING* AND N. H. MELCHIOR Bacteriological Research Department, Leo Pharmaceutical Products, DK-2750 Ballerup, Denmark Received for publication 26 December 1974 A newly described 6d-amidinopenicillanic acid derivative, mecillinam (for- merly called FL 1060), showed a high in vitro activity against Enterobacteriac- eae. The effect on Escherichia coli was bactericidal and was due to lysis of the cells. The longer the culture grew under the influence of mecillinam or the lower the inoculum, the greater the bactericidal effect. The morphology of the cells changed towards large spheric forms (2 to 5 ,m) under the influence of mecillinam. Consequently a great discrepancy between the optical density and the viable count was seen. The morphologically abnormal cells could be protected against lysis in vitro by addition of ionized compounds such as sodium chloride. Abnormal cells were more sensitive to ampicillin than normal cells. As expected synergy could be demonstrated between mecillinam and ampicillin. This was marked under experimental conditions where the abnormal cells were protected against lysis. In a previous paper from this laboratory a new the liquid media was measured by the cryostatic group of penicillanic acid derivatives, 6,- method using a Knauer type M osmometer, and the amidinopenicillanic acids, with unusual in specific conductivity was measured as millisiemens at vitro antibacterial properties was described (4). 36 C using a conductivity meter, Radiometer type One member of CDM 2c. -

Ampicillin (Ampicillin Sodium) INJECTION, POWDER, FOR
Ampicillin for Injection, USP Rx Only (For Intramuscular or Intravenous Injection) To reduce the development of drug-resistant bacteria and maintain the effectiveness of ampicillin and other antibacterial drugs, ampicillin should be used only to treat or prevent infections that are proven or strongly suspected to be caused by bacteria. DESCRIPTION Ampicillin for injection, USP the monosodium salt of [2S-[2α,5α,6β(S*)]]-6- [(aminophenylacetyl)amino]-3,3-dimethyl-7-oxo-4-thia-1-azabicyclo[3.2.0]heptane-2-carboxylic acid, is a synthetic penicillin. It is an antibacterial agent with a broad spectrum of bactericidal activity against both penicillin-susceptible Gram-positive organisms and many common Gram-negative pathogens. Ampicillin for injection, USP is a white to cream-tinged, crystalline powder. The reconstituted solution is clear, colorless and free from visible particulates. Each vial of Ampicillin for injection, USP contains ampicillin sodium equivalent to 250 mg, 500 mg, 1 gram or 2 grams ampicillin. Ampicillin for injection, USP contains 65.8 mg [2.9 mEq] sodium per gram ampicillin. It has the following molecular structure: The molecular formula is C16H18N3NaO4S, and the molecular weight is 371.39. The pH range of the reconstituted solution is 8 to 10. CLINICAL PHARMACOLOGY Ampicillin for injection diffuses readily into most body tissues and fluids. However, penetration into the cerebrospinal fluid and brain occurs only when the meninges are inflamed. Ampicillin is excreted largely unchanged in the urine and its excretion can be delayed by concurrent administration of probenecid. Due to maturational changes in renal function, ampicillin half-life decreases as postmenstrual age (a sum of gestational age and postnatal age) increases for infants with postnatal age of less than 28 days. -

Australian Public Assessment Refport for Ceftaroline Fosamil (Zinforo)
Australian Public Assessment Report for ceftaroline fosamil Proprietary Product Name: Zinforo Sponsor: AstraZeneca Pty Ltd May 2013 Therapeutic Goods Administration About the Therapeutic Goods Administration (TGA) • The Therapeutic Goods Administration (TGA) is part of the Australian Government Department of Health and Ageing, and is responsible for regulating medicines and medical devices. • The TGA administers the Therapeutic Goods Act 1989 (the Act), applying a risk management approach designed to ensure therapeutic goods supplied in Australia meet acceptable standards of quality, safety and efficacy (performance), when necessary. • The work of the TGA is based on applying scientific and clinical expertise to decision- making, to ensure that the benefits to consumers outweigh any risks associated with the use of medicines and medical devices. • The TGA relies on the public, healthcare professionals and industry to report problems with medicines or medical devices. TGA investigates reports received by it to determine any necessary regulatory action. • To report a problem with a medicine or medical device, please see the information on the TGA website <http://www.tga.gov.au>. About AusPARs • An Australian Public Assessment Record (AusPAR) provides information about the evaluation of a prescription medicine and the considerations that led the TGA to approve or not approve a prescription medicine submission. • AusPARs are prepared and published by the TGA. • An AusPAR is prepared for submissions that relate to new chemical entities, generic medicines, major variations, and extensions of indications. • An AusPAR is a static document, in that it will provide information that relates to a submission at a particular point in time. • A new AusPAR will be developed to reflect changes to indications and/or major variations to a prescription medicine subject to evaluation by the TGA. -

Comparative Ceftaroline Activity Tested Against Staphylococcus Aureus Associated with Acute Bacterial Skin and Skin Structure Infection from a Tertiary Care Center
RESEARCH PAPER Medical Science Volume : 5 | Issue : 4 | April 2015 | ISSN - 2249-555X Comparative Ceftaroline Activity Tested Against Staphylococcus Aureus Associated with Acute Bacterial Skin and Skin Structure Infection from A Tertiary Care Center KEYWORDS MRSA, Ceftaroline, vancomycin Dr. Umamageswari S. S. M. Dr. S. Habeeb Mohammed Dr. Shameem Banu Associate Professor- Microbiology Associate Professor-Surgery Professor and HOD- Microbiology Chettinad Hospital & Research Chettinad Hospital & Research Chettinad Hospital & Research Insitute Insitute Insitute Dr. Jayanthi S. Associate Professor- Microbiology Chettinad Hospital & Research Insitute ABSTRACT Methicillin resistant Staphylococcus aureus (MRSA), commonest cause of acute bacterial skin and skin structure infection (ABSSSI) has only limited treatment options like vancomycin, Linezolid and teicoplanin. Staphylococcus aureus with reduced susceptibilities to vancomycin is developing nowadays among nosocomial infec- tion. A newest cephalosporin – Ceftaroline has received FDA approval for the treatment of ABSSSI recently has unique activity against MRSA. The aim of the study is to know the prevalence of Staphylococcus aureus in ABSSSI patients and to evaluate the in vitro activity of Ceftaroline, in comparison with other drugs. Minimum Inhibitory Concentration for vancomycin and Ceftaroline were determined by E-test strips. Out of 235 Staphylococcus aureus isolated from ABSSSI patient 32% were MRSA. No vancomycin resistant strain isolated in our study. In vitro, Ceftaroline control the growth of MRSA very effectively at 2mg/ml itself. Ceftaroline is a most welcomed drug in the treatment of MRSA. INTRODUCTION: MATERIALS AND METHODS: Complicated skin and skin structure infection (CSSSIs), now This study was carried out in Chettinad health city and known by the new US FDA as Acute bacterial skin and skin Research Institute, a tertiary care center hospital (Chen- structure infections (ABSSSI) are the most common infec- nai) between April 2013 and May 2014. -

AMEG Categorisation of Antibiotics
12 December 2019 EMA/CVMP/CHMP/682198/2017 Committee for Medicinal Products for Veterinary use (CVMP) Committee for Medicinal Products for Human Use (CHMP) Categorisation of antibiotics in the European Union Answer to the request from the European Commission for updating the scientific advice on the impact on public health and animal health of the use of antibiotics in animals Agreed by the Antimicrobial Advice ad hoc Expert Group (AMEG) 29 October 2018 Adopted by the CVMP for release for consultation 24 January 2019 Adopted by the CHMP for release for consultation 31 January 2019 Start of public consultation 5 February 2019 End of consultation (deadline for comments) 30 April 2019 Agreed by the Antimicrobial Advice ad hoc Expert Group (AMEG) 19 November 2019 Adopted by the CVMP 5 December 2019 Adopted by the CHMP 12 December 2019 Official address Domenico Scarlattilaan 6 ● 1083 HS Amsterdam ● The Netherlands Address for visits and deliveries Refer to www.ema.europa.eu/how-to-find-us Send us a question Go to www.ema.europa.eu/contact Telephone +31 (0)88 781 6000 An agency of the European Union © European Medicines Agency, 2020. Reproduction is authorised provided the source is acknowledged. Categorisation of antibiotics in the European Union Table of Contents 1. Summary assessment and recommendations .......................................... 3 2. Introduction ............................................................................................ 7 2.1. Background ........................................................................................................ -

Penicillin Allergy Guidance Document
Penicillin Allergy Guidance Document Key Points Background Careful evaluation of antibiotic allergy and prior tolerance history is essential to providing optimal treatment The true incidence of penicillin hypersensitivity amongst patients in the United States is less than 1% Alterations in antibiotic prescribing due to reported penicillin allergy has been shown to result in higher costs, increased risk of antibiotic resistance, and worse patient outcomes Cross-reactivity between truly penicillin allergic patients and later generation cephalosporins and/or carbapenems is rare Evaluation of Penicillin Allergy Obtain a detailed history of allergic reaction Classify the type and severity of the reaction paying particular attention to any IgE-mediated reactions (e.g., anaphylaxis, hives, angioedema, etc.) (Table 1) Evaluate prior tolerance of beta-lactam antibiotics utilizing patient interview or the electronic medical record Recommendations for Challenging Penicillin Allergic Patients See Figure 1 Follow-Up Document tolerance or intolerance in the patient’s allergy history Consider referring to allergy clinic for skin testing Created July 2017 by Macey Wolfe, PharmD; John Schoen, PharmD, BCPS; Scott Bergman, PharmD, BCPS; Sara May, MD; and Trevor Van Schooneveld, MD, FACP Disclaimer: This resource is intended for non-commercial educational and quality improvement purposes. Outside entities may utilize for these purposes, but must acknowledge the source. The guidance is intended to assist practitioners in managing a clinical situation but is not mandatory. The interprofessional group of authors have made considerable efforts to ensure the information upon which they are based is accurate and up to date. Any treatments have some inherent risk. Recommendations are meant to improve quality of patient care yet should not replace clinical judgment.