HLF Is a Potential Prognostic Biomarker in Head and Neck Squamous Cell Carcinoma Based on Bioinformatic Analysis and Experimental Validation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplemental Materials ZNF281 Enhances Cardiac Reprogramming
Supplemental Materials ZNF281 enhances cardiac reprogramming by modulating cardiac and inflammatory gene expression Huanyu Zhou, Maria Gabriela Morales, Hisayuki Hashimoto, Matthew E. Dickson, Kunhua Song, Wenduo Ye, Min S. Kim, Hanspeter Niederstrasser, Zhaoning Wang, Beibei Chen, Bruce A. Posner, Rhonda Bassel-Duby and Eric N. Olson Supplemental Table 1; related to Figure 1. Supplemental Table 2; related to Figure 1. Supplemental Table 3; related to the “quantitative mRNA measurement” in Materials and Methods section. Supplemental Table 4; related to the “ChIP-seq, gene ontology and pathway analysis” and “RNA-seq” and gene ontology analysis” in Materials and Methods section. Supplemental Figure S1; related to Figure 1. Supplemental Figure S2; related to Figure 2. Supplemental Figure S3; related to Figure 3. Supplemental Figure S4; related to Figure 4. Supplemental Figure S5; related to Figure 6. Supplemental Table S1. Genes included in human retroviral ORF cDNA library. Gene Gene Gene Gene Gene Gene Gene Gene Symbol Symbol Symbol Symbol Symbol Symbol Symbol Symbol AATF BMP8A CEBPE CTNNB1 ESR2 GDF3 HOXA5 IL17D ADIPOQ BRPF1 CEBPG CUX1 ESRRA GDF6 HOXA6 IL17F ADNP BRPF3 CERS1 CX3CL1 ETS1 GIN1 HOXA7 IL18 AEBP1 BUD31 CERS2 CXCL10 ETS2 GLIS3 HOXB1 IL19 AFF4 C17ORF77 CERS4 CXCL11 ETV3 GMEB1 HOXB13 IL1A AHR C1QTNF4 CFL2 CXCL12 ETV7 GPBP1 HOXB5 IL1B AIMP1 C21ORF66 CHIA CXCL13 FAM3B GPER HOXB6 IL1F3 ALS2CR8 CBFA2T2 CIR1 CXCL14 FAM3D GPI HOXB7 IL1F5 ALX1 CBFA2T3 CITED1 CXCL16 FASLG GREM1 HOXB9 IL1F6 ARGFX CBFB CITED2 CXCL3 FBLN1 GREM2 HOXC4 IL1F7 -

1714 Gene Comprehensive Cancer Panel Enriched for Clinically Actionable Genes with Additional Biologically Relevant Genes 400-500X Average Coverage on Tumor
xO GENE PANEL 1714 gene comprehensive cancer panel enriched for clinically actionable genes with additional biologically relevant genes 400-500x average coverage on tumor Genes A-C Genes D-F Genes G-I Genes J-L AATK ATAD2B BTG1 CDH7 CREM DACH1 EPHA1 FES G6PC3 HGF IL18RAP JADE1 LMO1 ABCA1 ATF1 BTG2 CDK1 CRHR1 DACH2 EPHA2 FEV G6PD HIF1A IL1R1 JAK1 LMO2 ABCB1 ATM BTG3 CDK10 CRK DAXX EPHA3 FGF1 GAB1 HIF1AN IL1R2 JAK2 LMO7 ABCB11 ATR BTK CDK11A CRKL DBH EPHA4 FGF10 GAB2 HIST1H1E IL1RAP JAK3 LMTK2 ABCB4 ATRX BTRC CDK11B CRLF2 DCC EPHA5 FGF11 GABPA HIST1H3B IL20RA JARID2 LMTK3 ABCC1 AURKA BUB1 CDK12 CRTC1 DCUN1D1 EPHA6 FGF12 GALNT12 HIST1H4E IL20RB JAZF1 LPHN2 ABCC2 AURKB BUB1B CDK13 CRTC2 DCUN1D2 EPHA7 FGF13 GATA1 HLA-A IL21R JMJD1C LPHN3 ABCG1 AURKC BUB3 CDK14 CRTC3 DDB2 EPHA8 FGF14 GATA2 HLA-B IL22RA1 JMJD4 LPP ABCG2 AXIN1 C11orf30 CDK15 CSF1 DDIT3 EPHB1 FGF16 GATA3 HLF IL22RA2 JMJD6 LRP1B ABI1 AXIN2 CACNA1C CDK16 CSF1R DDR1 EPHB2 FGF17 GATA5 HLTF IL23R JMJD7 LRP5 ABL1 AXL CACNA1S CDK17 CSF2RA DDR2 EPHB3 FGF18 GATA6 HMGA1 IL2RA JMJD8 LRP6 ABL2 B2M CACNB2 CDK18 CSF2RB DDX3X EPHB4 FGF19 GDNF HMGA2 IL2RB JUN LRRK2 ACE BABAM1 CADM2 CDK19 CSF3R DDX5 EPHB6 FGF2 GFI1 HMGCR IL2RG JUNB LSM1 ACSL6 BACH1 CALR CDK2 CSK DDX6 EPOR FGF20 GFI1B HNF1A IL3 JUND LTK ACTA2 BACH2 CAMTA1 CDK20 CSNK1D DEK ERBB2 FGF21 GFRA4 HNF1B IL3RA JUP LYL1 ACTC1 BAG4 CAPRIN2 CDK3 CSNK1E DHFR ERBB3 FGF22 GGCX HNRNPA3 IL4R KAT2A LYN ACVR1 BAI3 CARD10 CDK4 CTCF DHH ERBB4 FGF23 GHR HOXA10 IL5RA KAT2B LZTR1 ACVR1B BAP1 CARD11 CDK5 CTCFL DIAPH1 ERCC1 FGF3 GID4 HOXA11 IL6R KAT5 ACVR2A -

C/Ebpα Is an Essential Collaborator in Hoxa9/Meis1-Mediated Leukemogenesis
C/EBPα is an essential collaborator in Hoxa9/Meis1-mediated leukemogenesis Cailin Collinsa, Jingya Wanga, Hongzhi Miaoa, Joel Bronsteina, Humaira Nawera, Tao Xua, Maria Figueroaa, Andrew G. Munteana, and Jay L. Hessa,b,1 aDepartment of Pathology, University of Michigan, Ann Arbor, MI 48109; and bIndiana University School of Medicine, Indianapolis, IN 46202 Edited* by Louis M. Staudt, National Institutes of Health, Bethesda, MD, and approved May 19, 2014 (received for review February 12, 2014) Homeobox A9 (HOXA9) is a homeodomain-containing transcrip- with Hoxa9. In addition, C/EBP recognition motifs are enriched tion factor that plays a key role in hematopoietic stem cell expan- at Hoxa9 binding sites. sion and is commonly deregulated in human acute leukemias. A C/EBPα is a basic leucine-zipper transcription factor that plays variety of upstream genetic alterations in acute myeloid leukemia a critical role in lineage commitment during hematopoietic dif- −/− (AML) lead to overexpression of HOXA9, almost always in associ- ferentiation (18). Whereas Cebpa mice show complete loss of ation with overexpression of its cofactor meis homeobox 1 (MEIS1). the granulocytic compartment, recent work shows that loss of α A wide range of data suggests that HOXA9 and MEIS1 play a syn- C/EBP in adult HSCs leads to both an increase in the number ergistic causative role in AML, although the molecular mechanisms of functional HSCs and an increase in their proliferative and leading to transformation by HOXA9 and MEIS1 remain elusive. In repopulating capacity (19, 20). Conversely, CEBPA overexpression can promote transdifferentiation of a variety of fibroblastic cells to this study, we identify CCAAT/enhancer binding protein alpha (C/ the myeloid lineage and can induce monocytic differentiation in EBPα) as a critical collaborator required for Hoxa9/Meis1-mediated α MLL-fusion protein-mediated leukemias (21, 22). -

The Role of Constitutive Androstane Receptor and Estrogen Sulfotransferase in Energy Homeostasis
THE ROLE OF CONSTITUTIVE ANDROSTANE RECEPTOR AND ESTROGEN SULFOTRANSFERASE IN ENERGY HOMEOSTASIS by Jie Gao Bachelor of Engineering, China Pharmaceutical University, 1999 Master of Science, China Pharmaceutical University, 2002 Submitted to the Graduate Faculty of School of Pharmacy in partial fulfillment of the requirements for the degree of Doctor of Philosophy University of Pittsburgh 2012 UNIVERSITY OF PITTSBURGH School of Pharmacy This dissertation was presented by Jie Gao It was defended on January 18, 2012 and approved by Billy W. Day, Professor, Pharmaceutical Sciences Donald B. DeFranco, Professor, Pharmacology & Chemical Biology Samuel M. Poloyac, Associate Professor, Pharmaceutical Sciences Song Li, Associate Professor, Pharmaceutical Sciences Dissertation Advisor: Wen Xie, Professor, Pharmaceutical Sciences ii Copyright © by Jie Gao 2012 iii THE ROLE OF CONSTITUTIVE ANDROSTANE RECEPTOR AND ESTROGEN SULFOTRANSFERASE IN ENERGY HOMEOSTASIS Jie Gao, PhD University of Pittsburgh, 2012 Obesity and type 2 diabetes are related metabolic disorders of high prevalence. The constitutive androstane receptor (CAR) was initially characterized as a xenobiotic receptor regulating the responses of mammals to xenotoxicants. In this study, I have uncovered an unexpected role of CAR in preventing obesity and alleviating type 2 diabetes. Activation of CAR prevented obesity and improved insulin sensitivity in both the HFD-induced type 2 diabetic model and the ob/ob mice. In contrast, CAR null mice maintained on a chow diet showed spontaneous insulin insensitivity. The metabolic benefits of CAR activation may have resulted from inhibition of hepatic lipogenesis and gluconeogenesis. The molecular mechanism through which CAR activation suppressed hepatic gluconeogenesis might be mediated via peroxisome proliferator- activated receptor gamma coactivator-1 alpha (PGC-1α). -
Drosophila and Human Transcriptomic Data Mining Provides Evidence for Therapeutic
Drosophila and human transcriptomic data mining provides evidence for therapeutic mechanism of pentylenetetrazole in Down syndrome Author Abhay Sharma Institute of Genomics and Integrative Biology Council of Scientific and Industrial Research Delhi University Campus, Mall Road Delhi 110007, India Tel: +91-11-27666156, Fax: +91-11-27662407 Email: [email protected] Nature Precedings : hdl:10101/npre.2010.4330.1 Posted 5 Apr 2010 Running head: Pentylenetetrazole mechanism in Down syndrome 1 Abstract Pentylenetetrazole (PTZ) has recently been found to ameliorate cognitive impairment in rodent models of Down syndrome (DS). The mechanism underlying PTZ’s therapeutic effect is however not clear. Microarray profiling has previously reported differential expression of genes in DS. No mammalian transcriptomic data on PTZ treatment however exists. Nevertheless, a Drosophila model inspired by rodent models of PTZ induced kindling plasticity has recently been described. Microarray profiling has shown PTZ’s downregulatory effect on gene expression in fly heads. In a comparative transcriptomics approach, I have analyzed the available microarray data in order to identify potential mechanism of PTZ action in DS. I find that transcriptomic correlates of chronic PTZ in Drosophila and DS counteract each other. A significant enrichment is observed between PTZ downregulated and DS upregulated genes, and a significant depletion between PTZ downregulated and DS dowwnregulated genes. Further, the common genes in PTZ Nature Precedings : hdl:10101/npre.2010.4330.1 Posted 5 Apr 2010 downregulated and DS upregulated sets show enrichment for MAP kinase pathway. My analysis suggests that downregulation of MAP kinase pathway may mediate therapeutic effect of PTZ in DS. Existing evidence implicating MAP kinase pathway in DS supports this observation. -

Mapping Mammalian Cell-Type-Specific Transcriptional
ORIGINAL RESEARCH published: 18 November 2015 doi: 10.3389/fgene.2015.00331 Mapping Mammalian Cell-type-specific Transcriptional Regulatory Networks Using KD-CAGE and ChIP-seq Data in the TC-YIK Cell Line Marina Lizio 1, 2, Yuri Ishizu 1, 2, Masayoshi Itoh 1, 2, 3, Timo Lassmann 1, 2, 4, Akira Hasegawa 1, 2, Atsutaka Kubosaki 1, Jessica Severin 1, 2, Hideya Kawaji 1, 2, 3, Yukio Nakamura 5, the FANTOM consortium 1, Harukazu Suzuki 1, 2, Yoshihide Hayashizaki 1, 3, Piero Carninci 1, 2 and Alistair R. R. Forrest 1, 2, 6* 1 RIKEN Center for Life Science Technologies, Yokohama, Japan, 2 Division of Genomic Technologies, RIKEN Center for Life Science Technologies, Yokohama, Japan, 3 RIKEN Preventive Medicine and Diagnosis Innovation Program, Yokohama, Edited by: Japan, 4 Telethon Kids Institute, The University of Western Australia, Subiaco, WA, Australia, 5 Cell Engineering Division, Edgar Wingender, RIKEN BioResource Center, Ibaraki, Japan, 6 QEII Medical Centre and Centre for Medical Research, Harry Perkins Institute of The University Medical Center Medical Research, The University of Western Australia, Nedlands, WA, Australia Göttingen, Germany Reviewed by: Mikael Boden, Mammals are composed of hundreds of different cell types with specialized functions. The University of Queensland, Each of these cellular phenotypes are controlled by different combinations of Australia transcription factors. Using a human non islet cell insulinoma cell line (TC-YIK) which Ka-Chun Wong, City University of Hong Kong, China expresses insulin and the majority of known pancreatic beta cell specific genes *Correspondence: as an example, we describe a general approach to identify key cell-type-specific Alistair R. -

The Rat Hepatic Leukemia Factor (HLF) Gene Encodes Two Transcriptional Activators with Distinct Circadian Rhythms, Tissue Distributions and Target Preferences
The EMBO Journal vol.14 no.17 pp.4307-4317, 1995 The rat hepatic leukemia factor (HLF) gene encodes two transcriptional activators with distinct circadian rhythms, tissue distributions and target preferences E.Falvey, F.Fleury-Olela and U.Schibler1 the putative chicken TEF ortholog (Iyer et al., 1991). All three factors, DBP, HLF and TEF, accumulate in liver cell Department of Molecular Biology, University of Geneva, nuclei (Mueller et al., 1990; P.Fonjallaz and U.Schibler, 30, Quai Ernest Ansermet, CH-1211 Geneva, Switzerland in preparation) and may therefore participate in hepatocyte- 'Corresponding author specific gene expression. A hallmark of DBP is its robust circadian expression pattern in the liver, where DBP RNA Hepatic leukemia factor (HLF) is a member of the and protein accumulate to remarkably higher levels in the PAR family of transcription regulatory proteins. We evening than in the morning (Wuarin and Schibler, 1990), have characterized the rat HLF gene and studied its and thus may influence circadian variations in liver func- expression and activity. The rat HLF gene is tran- tion (Wuarin et al., 1992). scribed from two alternative promoters, a and 0, with PAR proteins comprise a subfamily of the basic leucine different circadian amplitudes and tissue specificities. zipper proteins (bZIP), a major family of transcription The a RNA isoforms produce a 43 kDa protein, HLF43, factors characterized by a positively charged a-helical abundant in brain, liver and kidney, like the previously region involved in DNA binding and an adjacent amphi- described human HLF RNA. The J RNA HLF isoforms pathic a-helical region that allows homodimer formation use a CUG codon to initiate translation of a novel or dimerization with other related bZIP proteins 36 kDa protein, HLF36, which is shorter at its N- (Kouzarides and Ziff, 1988; Landschulz et al., 1988). -

Xo PANEL DNA GENE LIST
xO PANEL DNA GENE LIST ~1700 gene comprehensive cancer panel enriched for clinically actionable genes with additional biologically relevant genes (at 400 -500x average coverage on tumor) Genes A-C Genes D-F Genes G-I Genes J-L AATK ATAD2B BTG1 CDH7 CREM DACH1 EPHA1 FES G6PC3 HGF IL18RAP JADE1 LMO1 ABCA1 ATF1 BTG2 CDK1 CRHR1 DACH2 EPHA2 FEV G6PD HIF1A IL1R1 JAK1 LMO2 ABCB1 ATM BTG3 CDK10 CRK DAXX EPHA3 FGF1 GAB1 HIF1AN IL1R2 JAK2 LMO7 ABCB11 ATR BTK CDK11A CRKL DBH EPHA4 FGF10 GAB2 HIST1H1E IL1RAP JAK3 LMTK2 ABCB4 ATRX BTRC CDK11B CRLF2 DCC EPHA5 FGF11 GABPA HIST1H3B IL20RA JARID2 LMTK3 ABCC1 AURKA BUB1 CDK12 CRTC1 DCUN1D1 EPHA6 FGF12 GALNT12 HIST1H4E IL20RB JAZF1 LPHN2 ABCC2 AURKB BUB1B CDK13 CRTC2 DCUN1D2 EPHA7 FGF13 GATA1 HLA-A IL21R JMJD1C LPHN3 ABCG1 AURKC BUB3 CDK14 CRTC3 DDB2 EPHA8 FGF14 GATA2 HLA-B IL22RA1 JMJD4 LPP ABCG2 AXIN1 C11orf30 CDK15 CSF1 DDIT3 EPHB1 FGF16 GATA3 HLF IL22RA2 JMJD6 LRP1B ABI1 AXIN2 CACNA1C CDK16 CSF1R DDR1 EPHB2 FGF17 GATA5 HLTF IL23R JMJD7 LRP5 ABL1 AXL CACNA1S CDK17 CSF2RA DDR2 EPHB3 FGF18 GATA6 HMGA1 IL2RA JMJD8 LRP6 ABL2 B2M CACNB2 CDK18 CSF2RB DDX3X EPHB4 FGF19 GDNF HMGA2 IL2RB JUN LRRK2 ACE BABAM1 CADM2 CDK19 CSF3R DDX5 EPHB6 FGF2 GFI1 HMGCR IL2RG JUNB LSM1 ACSL6 BACH1 CALR CDK2 CSK DDX6 EPOR FGF20 GFI1B HNF1A IL3 JUND LTK ACTA2 BACH2 CAMTA1 CDK20 CSNK1D DEK ERBB2 FGF21 GFRA4 HNF1B IL3RA JUP LYL1 ACTC1 BAG4 CAPRIN2 CDK3 CSNK1E DHFR ERBB3 FGF22 GGCX HNRNPA3 IL4R KAT2A LYN ACVR1 BAI3 CARD10 CDK4 CTCF DHH ERBB4 FGF23 GHR HOXA10 IL5RA KAT2B LZTR1 ACVR1B BAP1 CARD11 CDK5 CTCFL DIAPH1 ERCC1 FGF3 GID4 HOXA11 -

Genome-Wide Homozygosity Patterns and Evidence for Selection in a Set of European and Near Eastern Horse Breeds
Article Genome-Wide Homozygosity Patterns and Evidence for Selection in a Set of European and Near Eastern Horse Breeds Gertrud Grilz-Seger 1,*, Markus Neuditschko 2, Anne Ricard 3, Brandon Velie 4,5, Gabriella Lindgren 4,6,, Matjaz Mesarič 7, Marko Cotman 8, Michaela Horna 9, Max Dobretsberger 1, Gottfried Brem 1 and Thomas Druml 1 1 Institute of Animal Breeding and Genetics, University of Veterinary Sciences Vienna, Veterinärplatz 1, 1210 Vienna, Austria; [email protected] (M.D.); [email protected] (G.B.); [email protected] (T.D.) 2 Agroscope, Swiss National Stud Farm, Les Longs Prés, CH-1580 Avenches, Switzerland; [email protected] 3 UMR 1313 Génétique Animale et Biologie Intégrative, Institut National de la Recherche Agronomique, Domaine de Vilvert, Bat 211, 78352 Jouy-en-Josas, France; [email protected] 4 Department of Animal Breeding & Genetics, Swedish University of Agricultural Sciences, Ulls väg 26, 750 07 Uppsala, Sweden; [email protected] (B.V.); [email protected] (G.L.) 5 School of Life and Environmental Sciences, University of Sydney, Eastern Ave, 2006 NSW Sydney, Australia 6 Livestock Genetics, Department of Biosystems, KU Leuven, 3001 Leuven, Belgium 7 Clinic for Reproduction and Large Animals, University of Ljubljana, Veterinary, Faculty, Cesta v Mestni log 47, 1000 Ljubljana, Slovenia; [email protected] 8 Institute for Preclinical Sciences, University of Ljubljana, Veterinary Faculty, Gerbičeva 60, 1000 Ljubljana, Slovenia; [email protected] 9 Department of Animal Husbandry, Slovak University of Agriculture in Nitra, Tr. A. Hlinku 2, 949 76 Nitra, Slovakia; [email protected] * Correspondence: [email protected] Received: 14 May 2019; Accepted: 26 June 2019; Published: 28 June 2019 Abstract: Intensive artificial and natural selection have shaped substantial variation among European horse breeds. -

Integrated Analysis of Whole Genome and Transcriptome Sequencing in a Young Patient with Gastric Cancer Provides Insights for Precision Therapy
ONCOLOGY LETTERS 20: 115, 2020 Integrated analysis of whole genome and transcriptome sequencing in a young patient with gastric cancer provides insights for precision therapy KONGWANG HU1, WEIQIANG YU2, OLUGBENGA EMMANUEL AJAYI2, LONGLONG LI1, ZHIGUO HUANG1, QIQI RONG2, SHUAILI WANG2 and QING‑FA WU2 1Division of Gastrointestinal Surgery, Department of General Surgery, The First Affiliated Hospital of Anhui Medical University, Hefei, Anhui 230022; 2Hefei National Laboratory for Physical Sciences at Microscale, CAS Key Laboratory of Innate Immunity and Chronic Disease, School of Basic Medical Sciences, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, Anhui 230026, P.R. China Received June 4, 2019; Accepted July 17, 2020 DOI: 10.3892/ol.2020.11976 Abstract. Gastric cancer is a leading cause of cancer‑ MUC4, SLC8A2) mutations, or structural variations (CCND3, associated deaths worldwide and is considered to be an age‑related FGFR2 and FGFR3) were differentially expressed in the patient disease. In younger patients, gastric cancer is biologically more and could be promising precision therapy targets. aggressive, and prognosis is worse compared with that in elderly patients. In the present case report, the whole genome and tran‑ Introduction scriptome was sequenced in a 26‑year‑old patient with gastric cancer who presented with gastric cancer‑related symptoms and Gastric cancer (GC) is a leading cause of cancer‑related deaths was admitted to the First Affiliated Anhui Medical Hospital worldwide and is known to be an age‑related disease (1). (Hefei, China) in December 2016. In total, 9 germline and 4 The mean age of patients with GC at diagnosis is ~60 years somatic mutations were identified in the patient, and there were and <3% of GC cases are reported in patients <30 years of more deleterious sites in the germline mutated genes. -

Robles JTO Supplemental Digital Content 1
Supplementary Materials An Integrated Prognostic Classifier for Stage I Lung Adenocarcinoma based on mRNA, microRNA and DNA Methylation Biomarkers Ana I. Robles1, Eri Arai2, Ewy A. Mathé1, Hirokazu Okayama1, Aaron Schetter1, Derek Brown1, David Petersen3, Elise D. Bowman1, Rintaro Noro1, Judith A. Welsh1, Daniel C. Edelman3, Holly S. Stevenson3, Yonghong Wang3, Naoto Tsuchiya4, Takashi Kohno4, Vidar Skaug5, Steen Mollerup5, Aage Haugen5, Paul S. Meltzer3, Jun Yokota6, Yae Kanai2 and Curtis C. Harris1 Affiliations: 1Laboratory of Human Carcinogenesis, NCI-CCR, National Institutes of Health, Bethesda, MD 20892, USA. 2Division of Molecular Pathology, National Cancer Center Research Institute, Tokyo 104-0045, Japan. 3Genetics Branch, NCI-CCR, National Institutes of Health, Bethesda, MD 20892, USA. 4Division of Genome Biology, National Cancer Center Research Institute, Tokyo 104-0045, Japan. 5Department of Chemical and Biological Working Environment, National Institute of Occupational Health, NO-0033 Oslo, Norway. 6Genomics and Epigenomics of Cancer Prediction Program, Institute of Predictive and Personalized Medicine of Cancer (IMPPC), 08916 Badalona (Barcelona), Spain. List of Supplementary Materials Supplementary Materials and Methods Fig. S1. Hierarchical clustering of based on CpG sites differentially-methylated in Stage I ADC compared to non-tumor adjacent tissues. Fig. S2. Confirmatory pyrosequencing analysis of DNA methylation at the HOXA9 locus in Stage I ADC from a subset of the NCI microarray cohort. 1 Fig. S3. Methylation Beta-values for HOXA9 probe cg26521404 in Stage I ADC samples from Japan. Fig. S4. Kaplan-Meier analysis of HOXA9 promoter methylation in a published cohort of Stage I lung ADC (J Clin Oncol 2013;31(32):4140-7). Fig. S5. Kaplan-Meier analysis of a combined prognostic biomarker in Stage I lung ADC. -
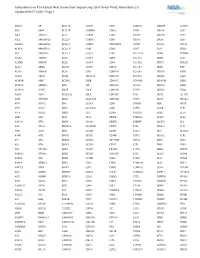
Comprehensive Pan-Cancer Next Generation Sequencing Solid Tumor Panel, Aberration List Updated 08-07-2020 --Page 1
Comprehensive Pan-Cancer Next Generation Sequencing Solid Tumor Panel, Aberration List Updated 08-07-2020 --Page 1 ABCC3 AR BCL11A CANT1 CDK1 CMKLR1 DAB2IP DUSP9 ABI1 ARAF BCL11B CAPRIN1 CDK12 CNBP DACH1 E2F1 ABL1 ARFRP1 BCL2 CAPZB CDK2 CNOT2 DACH2 E2F3 ABL2 ARHGAP20 BCL2A1 CARD11 CDK4 CNTN1 DAXX EBF1 ABLIM1 ARHGAP26 BCL2L1 CARM1 CDK5RAP2 CNTRL DCLK2 ECT2L ACACA ARHGEF12 BCL2L11 CARS CDK6 COG5 DCN EDIL3 ACE ARHGEF7 BCL2L2 CASC5 CDK7 COL11A1 DDB1 EDNRB ACER1 ARID1A BCL3 CASP3 CDK8 COL1A1 DDB2 EED ACSBG1 ARID1B BCL6 CASP7 CDK9 COL1A2 DDIT3 EEFSEC ACSL3 ARID2 BCL7A CASP8 CDKL5 COL3A1 DDR2 EGF ACSL6 ARID5B BCL9 CAV1 CDKN1A COL6A3 DDX10 EGFR ACVR1 ARIH2 BCOR CBFA2T3 CDKN1B COL9A3 DDX20 EGR1 ACVR1B ARNT BCORL1 CBFB CDKN1C COMMD1 DDX39B EGR2 ACVR1C ARRDC4 BCR CBL CDKN2A COX6C DDX3X EGR3 ACVR2A ASMTL BDNF CBLB CDKN2B CPNE1 DDX41 EGR4 ADD3 ASPH BHLHE22 CBLC CDKN2C CPS1 DDX5 EIF1AX ADM ASPSCR1 BICC1 CCDC28A CDKN2D CPSF6 DDX6 EIF4A2 AFF1 ASTN2 BIN1 CCDC6 CDX1 CRADD DEK EIF4E AFF3 ASXL1 BIRC3 CCDC88C CDX2 CREB1 DGKB ELF3 AFF4 ASXL2 BIRC6 CCK CEBPA CREB3L1 DGKI ELF4 AGR3 ATF1 BLM CCL2 CEBPB CREB3L2 DGKZ ELK4 AHCYL1 ATF3 BMP4 CCNA2 CEBPD CREBBP DICER1 ELL AHI1 ATG13 BMPR1A CCNB1IP1 CEBPE CRKL DIRAS3 ELN AHR ATG5 BRAF CCNB3 CENPF CRLF2 DIS3 ELOVL2 AHRR ATIC BRCA1 CCND1 CENPU CRTC1 DIS3L2 ELP2 AIP ATL1 BRCA2 CCND2 CEP170B CRTC3 DKK1 EML1 AK2 ATM BRCC3 CCND3 CEP57 CSF1 DKK2 EML4 AK5 ATP1B4 BRD1 CCNE1 CEP85L CSF1R DKK4 ENPP2 AKAP12 ATP8A2 BRD3 CCNG1 CHCHD7 CSF3 DLEC1 EP300 AKAP6 ATR BRD4 CCT6B CHD2 CSF3R DLL1 EP400 AKAP9 ATRNL1 BRIP1 CD19 CHD4 CSNK1A1 DLL3