Fusiforma Themisticola N. Gen., N. Sp., a New Genus and Species Of
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

ABSTRACT Gregarine Parasitism in Dragonfly Populations of Central
ABSTRACT Gregarine Parasitism in Dragonfly Populations of Central Texas with an Assessment of Fitness Costs in Erythemis simplicicollis Jason L. Locklin, Ph.D. Mentor: Darrell S. Vodopich, Ph.D. Dragonfly parasites are widespread and frequently include gregarines (Phylum Apicomplexa) in the gut of the host. Gregarines are ubiquitous protozoan parasites that infect arthropods worldwide. More than 1,600 gregarine species have been described, but only a small percentage of invertebrates have been surveyed for these apicomplexan parasites. Some consider gregarines rather harmless, but recent studies suggest otherwise. Odonate-gregarine studies have more commonly involved damselflies, and some have considered gregarines to rarely infect dragonflies. In this study, dragonfly populations were surveyed for gregarines and an assessment of fitness costs was made in a common and widespread host species, Erythemis simplicicollis. Adult dragonfly populations were surveyed weekly at two reservoirs in close proximity to one another and at a flow-through wetland system. Gregarine prevalences and intensities were compared within host populations between genders, among locations, among wing loads, and through time. Host fitness parameters measured included wing load, egg size, clutch size, and total egg count. Of the 37 dragonfly species surveyed, 14 species (38%) hosted gregarines. Thirteen of those species were previously unreported as hosts. Gregarine prevalences ranged from 2% – 52%. Intensities ranged from 1 – 201. Parasites were aggregated among their hosts. Gregarines were found only in individuals exceeding a minimum wing load, indicating that gregarines are likely not transferred from the naiad to adult during emergence. Prevalence and intensity exhibited strong seasonality during both years at one of the reservoirs, but no seasonal trend was detected at the wetland. -

Special Issue Featuring: a Case Study on Black Gill in Georgia Shrimp
Volume 29 • Number 4 • Winter 2015 Special Issue Featuring: A Case Study on Black Gill in Georgia Shrimp Volume 29 • Number 4 • Winter 2015 The Mystery of Black Gill: Shrimpers in the South Atlantic Face Off with a Cryptic Parasite BY JILL M. GAMBILL, ALLISON E. DOYLE, RICHARD F. LEE, PH.D., PATRICK J. GEER, ANNA N. WALKER, PH.D., LINDSEY G. PARKER, PH.D., AND MARC E. FRISCHER, PH.D. ABSTRACT In the Southeast United States, an unidentified parasite is infecting shrimp and presenting new challenges for an already struggling industry. Emerging research in Georgia is investigating the resulting condition, known as Black Gill, to better understand this newest threat to the state’s most valuable commercial fishery. Researchers, shrimpers, extension agents, and fishery managers are working collaboratively to gather baseline data on where, when, and how frequently Black Gill is occurring, as well as partnering to determine its epidemiology, dispersal, and possible intervention strategies. Savannah, Ga. – In 2013, after years of intense competition from lower priced imports and financial pressures stemming Figure 1. A Georgia white shrimp displays symptoms of the from the rising cost of fuel and insurance, Georgia shrimpers Black Gill condition around its gills. Courtesy of Rachael were poised for a comeback as they looked forward to a Randall and Chelsea Parrish, 2015 profitable year. Shrimp prices had tripled, as a consequence of a bacterial disease infecting the supply of farmed shrimp A LANDMARK YEAR from Asia (Loc et al. 2013). Commercial food shrimp landings in 2013 were the lowest Georgia shrimpers took to the water with high expectations, in recent history. -

First Report of Some Trichodinid Cilioph (Ciliophora: Trichodinidae
SpeciesREPORT, Vol. 17A,RTICLE No. 56, July 1, 2016 REPORT ISSN 2319–5746 EISSN 2319–5754 SpeciesAn International Journal First report of some Trichodinid Ciliophorans (Ciliophora: Peritrichida) (Ciliophora: Trichodinidae) parasitizing cultured Oranda Gold Fish (Carassius auratus auratus L.) in India Saha M1, Mondal S2, Mandal SK3, Mitra P4, Das K5 and Bandyopadhyay PK6☼ ( 1-6) Parasitology Laboratory, Department of Zoology, University of Kalyani, Kalyani 741235, West Bengal, India ☼Corresponding author : Bandyopadhyay PK, Parasitology Laboratory, Department of Zoology, University of Kalyani, Kalyani 741235, West Bengal, India. Publication History Received: 7 August 2016 Accepted: 28 August 2016 Online First: 1 September 2016 Published: July-September 2016 Citation Saha M, Mondal S, Mandal SK, Mitra P, Das K, Bandyopadhyay PK. First report of some Trichodinid Ciliophorans (Ciliophora: Peritrichida) (Ciliophora: Trichodinidae) parasitizing cultured Oranda Gold Fish (Carassius auratus auratus L.) in India. Species, 2016, 17(56), 131-140 Publication License This work is licensed under a Creative Commons Attribution 4.0 International License. General Note Article is recommended to print as color digital color version in recycled paper. ABSTRACT Ornamental fish culture considered as one of the most important means of home entertainment, because of its diversity and beauty of picturesque colors. Trichodiniasis of fishes causing harm and economic losses in this fish industry. There are many records of trichodinids ectoparasites infesting fish have been found throughout the World, but no such study has been conducted on this 131 131 131 parasitic group infesting ornamental fish Carassius auratus auratus L. in India. During the survey, four already known trichodinid Saha et al. Page Page Page First report of some Trichodinid Ciliophorans (Ciliophora: Peritrichida) (Ciliophora: Trichodinidae) parasitizing cultured Oranda Gold Fish (Carassius auratus auratus L.) in India, Species, 2016, 17(56), 131-140, www.discoveryjournals.com © 2016 Discovery Publication. -

Penaeid Shrimp in Chesapeake Bay: Population Growth and Black Gill Disease Syndrome
W&M ScholarWorks VIMS Articles Virginia Institute of Marine Science 2021 Penaeid Shrimp in Chesapeake Bay: Population Growth and Black Gill Disease Syndrome Troy D. Tuckey Virginia Institute of Marine Science Jillian L. Swinford Mary C. Fabrizio Virginia Institute of Marine Science Hamish J. Small Virginia Institute of Marine Science Jeffrey D. Shields Virginia Institute of Marine Science Follow this and additional works at: https://scholarworks.wm.edu/vimsarticles Part of the Aquaculture and Fisheries Commons, and the Marine Biology Commons Recommended Citation Tuckey, Troy D.; Swinford, Jillian L.; Fabrizio, Mary C.; Small, Hamish J.; and Shields, Jeffrey D., Penaeid Shrimp in Chesapeake Bay: Population Growth and Black Gill Disease Syndrome (2021). Marine and Coastal Fisheries, 13, 159-173. DOI: 10.1002/mcf2.10143 This Article is brought to you for free and open access by the Virginia Institute of Marine Science at W&M ScholarWorks. It has been accepted for inclusion in VIMS Articles by an authorized administrator of W&M ScholarWorks. For more information, please contact [email protected]. Marine and Coastal Fisheries 13:159–173, 2021 © 2021 The Authors. Marine and Coastal Fisheries published by Wiley Periodicals LLC on behalf of American Fisheries Society ISSN: 1942-5120 online DOI: 10.1002/mcf2.10143 ARTICLE Penaeid Shrimp in Chesapeake Bay: Population Growth and Black Gill Disease Syndrome Troy D. Tuckey* Virginia Institute of Marine Science, William & Mary, 1370 Greate Road, Gloucester Point, Virginia 23062, USA Jillian L. Swinford Texas Parks and Wildlife, Coastal Fisheries Division, Perry R. Bass Marine Fisheries Research Center, 3864 Farm to Market Road 3280, Palacios, Texas 77465, USA Mary C. -
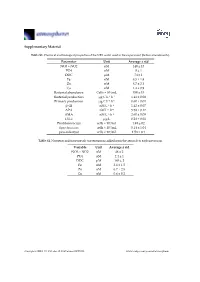
Supplementary Material Parameter Unit Average ± Std NO3 + NO2 Nm
Supplementary Material Table S1. Chemical and biological properties of the NRS water used in the experiment (before amendments). Parameter Unit Average ± std NO3 + NO2 nM 140 ± 13 PO4 nM 8 ± 1 DOC μM 74 ± 1 Fe nM 8.5 ± 1.8 Zn nM 8.7 ± 2.1 Cu nM 1.4 ± 0.9 Bacterial abundance Cells × 104/mL 350 ± 15 Bacterial production μg C L−1 h−1 1.41 ± 0.08 Primary production μg C L−1 h−1 0.60 ± 0.01 β-Gl nM L−1 h−1 1.42 ± 0.07 APA nM L−1 h−1 5.58 ± 0.17 AMA nM L−1·h−1 2.60 ± 0.09 Chl-a μg/L 0.28 ± 0.01 Prochlorococcus cells × 104/mL 1.49 ± 02 Synechococcus cells × 104/mL 5.14 ± 1.04 pico-eukaryot cells × 103/mL 1.58 × 0.1 Table S2. Nutrients and trace metals concentrations added from the aerosols to each mesocosm. Variable Unit Average ± std NO3 + NO2 nM 48 ± 2 PO4 nM 2.4 ± 1 DOC μM 165 ± 2 Fe nM 2.6 ± 1.5 Zn nM 6.7 ± 2.5 Cu nM 0.6 ± 0.2 Atmosphere 2019, 10, 358; doi:10.3390/atmos10070358 www.mdpi.com/journal/atmosphere Atmosphere 2019, 10, 358 2 of 6 Table S3. ANOVA test results between control, ‘UV-treated’ and ‘live-dust’ treatments at 20 h or 44 h, with significantly different values shown in bold. ANOVA df Sum Sq Mean Sq F Value p-value Chl-a 20 H 2, 6 0.03, 0.02 0.02, 0 4.52 0.0634 44 H 2, 6 0.02, 0 0.01, 0 23.13 0.002 Synechococcus Abundance 20 H 2, 7 8.23 × 107, 4.11 × 107 4.11 × 107, 4.51 × 107 0.91 0.4509 44 H 2, 7 5.31 × 108, 6.97 × 107 2.65 × 108, 1.16 × 107 22.84 0.0016 Prochlorococcus Abundance 20 H 2, 8 4.22 × 107, 2.11 × 107 2.11 × 107, 2.71 × 106 7.77 0.0216 44 H 2, 8 9.02 × 107, 1.47 × 107 4.51 × 107, 2.45 × 106 18.38 0.0028 Pico-eukaryote -

Supplementary Figure 1 Multicenter Randomised Control Trial 2746 Randomised
Supplementary Figure 1 Multicenter randomised control trial 2746 randomised 947 control 910 MNP without zinc 889 MNP with zinc 223 lost to follow up 219 lost to follow up 183 lost to follow up 34 refused 29 refused 37 refused 16 died 12 died 9 died 3 excluded 4 excluded 1 excluded 671 in follow-up 646 in follow-up 659 in follow-up at 24mo of age at 24mo of age at 24mo of age Selection for Microbiome sequencing 516 paired samples unavailable 469 paired samples unavailable 497 paired samples unavailable 69 antibiotic use 63 antibiotic use 67 antibiotic use 31 outside of WLZ criteria 37 outside of WLZ criteria 34 outside of WLZ criteria 6 diarrhea last 7 days 2 diarrhea last 7 days 7 diarrhea last 7 days 39 WLZ > -1 at 24 mo 10 WLZ < -2 at 24mo 58 WLZ > -1 at 24 mo 17 WLZ < -2 at 24mo 48 WLZ > -1 at 24 mo 8 WLZ < -2 at 24mo available for selection available for selection available for selection available for selection available for selection1 available for selection1 14 selected 10 selected 15 selected 14 selected 20 selected1 7 selected1 1 Two subjects (one in the reference WLZ group and one undernourished) had, at 12 months, no diarrhea within 1 day of stool collection but reported diarrhea within 7 days prior. Length, cm kg Weight, Supplementary Figure 2. Length (left) and weight (right) z-scores of children recruited into clinical trial NCT00705445 during the first 24 months of life. Median and quantile values are shown, with medians for participants profiled in current study indicated by red (undernourished) and black (reference WLZ) lines. -

Why the –Omic Future of Apicomplexa Should Include Gregarines Julie Boisard, Isabelle Florent
Why the –omic future of Apicomplexa should include Gregarines Julie Boisard, Isabelle Florent To cite this version: Julie Boisard, Isabelle Florent. Why the –omic future of Apicomplexa should include Gregarines. Biology of the Cell, Wiley, 2020, 10.1111/boc.202000006. hal-02553206 HAL Id: hal-02553206 https://hal.archives-ouvertes.fr/hal-02553206 Submitted on 24 Apr 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Article title: Why the –omic future of Apicomplexa should include Gregarines. Names of authors: Julie BOISARD1,2 and Isabelle FLORENT1 Authors affiliations: 1. Molécules de Communication et Adaptation des Microorganismes (MCAM, UMR 7245), Département Adaptations du Vivant (AVIV), Muséum National d’Histoire Naturelle, CNRS, CP52, 57 rue Cuvier 75231 Paris Cedex 05, France. 2. Structure et instabilité des génomes (STRING UMR 7196 CNRS / INSERM U1154), Département Adaptations du vivant (AVIV), Muséum National d'Histoire Naturelle, CP 26, 57 rue Cuvier 75231 Paris Cedex 05, France. Short Title: Gregarines –omics for Apicomplexa studies -

Prevalent Ciliate Symbiosis on Copepods: High Genetic Diversity and Wide Distribution Detected Using Small Subunit Ribosomal RNA Gene
Prevalent Ciliate Symbiosis on Copepods: High Genetic Diversity and Wide Distribution Detected Using Small Subunit Ribosomal RNA Gene Zhiling Guo1,2, Sheng Liu1, Simin Hu1, Tao Li1, Yousong Huang4, Guangxing Liu4, Huan Zhang2,4*, Senjie Lin2,3* 1 Key Laboratory of Marine Bio-resources Sustainable Utilization, South China Sea Institute of Oceanology, Chinese Academy of Science, Guangzhou, Guangdong, China, 2 Department of Marine Sciences, University of Connecticut, Groton, Connecticut, United States of America, 3 Marine Biodiversity and Global Change Laboratory, Xiamen University, Xiamen, Fujian, China, 4 Department of Environmental Science, Ocean University of China, Qingdao, Shandong, China Abstract Toward understanding the genetic diversity and distribution of copepod-associated symbiotic ciliates and the evolutionary relationships with their hosts in the marine environment, we developed a small subunit ribosomal RNA gene (18S rDNA)- based molecular method and investigated the genetic diversity and genotype distribution of the symbiotic ciliates on copepods. Of the 10 copepod species representing six families collected from six locations of Pacific and Atlantic Oceans, 9 were found to harbor ciliate symbionts. Phylogenetic analysis of the 391 ciliate 18S rDNA sequences obtained revealed seven groups (ribogroups), six (containing 99% of all the sequences) belonging to subclass Apostomatida, the other clustered with peritrich ciliate Vorticella gracilis. Among the Apostomatida groups, Group III were essentially identical to Vampyrophrya pelagica, and the other five groups represented the undocumented ciliates that were close to Vampyrophrya/ Gymnodinioides/Hyalophysa. Group VI ciliates were found in all copepod species but one (Calanus sinicus), and were most abundant among all ciliate sequences obtained, indicating that they are the dominant symbiotic ciliates universally associated with copepods. -

The Amoeboid Parabasalid Flagellate Gigantomonas Herculeaof
Acta Protozool. (2005) 44: 189 - 199 The Amoeboid Parabasalid Flagellate Gigantomonas herculea of the African Termite Hodotermes mossambicus Reinvestigated Using Immunological and Ultrastructural Techniques Guy BRUGEROLLE Biologie des Protistes, UMR 6023, CNRS and Université Blaise Pascal de Clermont-Ferrand, Aubière Cedex, France Summary. The amoeboid form of Gigantomonas herculea (Dogiel 1916, Kirby 1946), a symbiotic flagellate of the grass-eating subterranean termite Hodotermes mossambicus from East Africa, is observed by light, immunofluorescence and transmission electron microscopy. Amoeboid cells display a hyaline margin and a central granular area containing the nucleus, the internalized flagellar apparatus, and organelles such as Golgi bodies, hydrogenosomes, and food vacuoles with bacteria or wood particles. Immunofluorescence microscopy using monoclonal antibodies raised against Trichomonas vaginalis cytoskeleton, such as the anti-tubulin IG10, reveals the three long anteriorly-directed flagella, and the axostyle folded into the cytoplasm. A second antibody, 4E5, decorates the conspicuous crescent-shaped structure or cresta bordered by the adhering recurrent flagellum. Transmission electron micrographs show a microfibrillar network in the cytoplasmic margin and internal bundles of microfilaments similar to those of lobose amoebae that are indicative of cytoplasmic streaming. They also confirm the internalization of the flagella. The arrangement of basal bodies and fibre appendages, and the axostyle composed of a rolled sheet of microtubules are very close to that of the devescovinids Foaina and Devescovina. The very large microfibrillar cresta supporting an enlarged recurrent flagellum resembles that of Macrotrichomonas. The parabasal apparatus attached to the basal bodies is small in comparison to the cell size; this is probably related to the presence of many Golgi bodies supported by a striated fibre that are spread throughout the central cytoplasm in a similar way to Placojoenia and Mixotricha. -

Full-Text (PDF)
African Journal of Microbiology Research Vol. 6(9), pp. 2145-2149, 9 March, 2012 Available online at http://www.academicjournals.org/AJMR DOI: 10.5897/AJMR11.1582 ISSN 1996-0808 ©2012 Academic Journals Full Length Research Paper Two trichodinids of Paratrichodina (Ciliophora, Peritrichida, Trichodinidae) infecting gills of Ietalurus punetaus from Chongqing, China Fahui Tang, Yuanjun Zhao* and Chunning Liu Chongqing Key Laboratory of Animal Biology, Chongqing Normal University, Chongqing 400047, P. R. China. Accepted 13 January, 2012 During a parasitological survey in China, two species of trichodinid ectoparasite (Ciliophora, Peritrichida, Trichodinidae) belonging to genus Paratrichodina , were isolated from letalurus punetaus (Siluridae ). One species was identified as a new species and named as Paratrichodina rotundiformis sp. nov. and this new species has round and undeveloped blade with round tangent point, undeveloped triangular central part, slim ray with inclining backward, and whole fitting loose denticle. The other species, Paratrichodina corlissi is described for the first time in Asia in the present work. The morphometric data were obtained from specimens prepared using dry silver nitrate and methyl green- pyronin stain techniques. Results of comparison between the new species and closely related species are provided. Key words: Ciliophora, oligohymenophorea, Peritrichida, Paratrichodina , new species, Ietalurus punetaus. INTRODUCTION Trichodinid ciliates are well-known ectoparasites of MATERIALS AND METHODS fishes, amphibians -

Redalyc.Paratrichodina Africana (Ciliophora: Trichodinidae) of Wild and Cultured Nile Tilapia in the Northern Brazil
Revista Brasileira de Parasitologia Veterinária ISSN: 0103-846X [email protected] Colégio Brasileiro de Parasitologia Veterinária Brasil Tavares-Dias, Marcos; da Costa Marchiori, Natália; Laterça Martins, Maurício Paratrichodina africana (Ciliophora: Trichodinidae) of wild and cultured Nile tilapia in the Northern Brazil Revista Brasileira de Parasitologia Veterinária, vol. 22, núm. 2, abril-junio, 2013, pp. 248- 252 Colégio Brasileiro de Parasitologia Veterinária Jaboticabal, Brasil Available in: http://www.redalyc.org/articulo.oa?id=397841488011 How to cite Complete issue Scientific Information System More information about this article Network of Scientific Journals from Latin America, the Caribbean, Spain and Portugal Journal's homepage in redalyc.org Non-profit academic project, developed under the open access initiative Full Article Rev. Bras. Parasitol. Vet., Jaboticabal, v. 22, n. 2, p. 248-252, abr.-jun. 2013 ISSN 0103-846X (impresso) / ISSN 1984-2961 (eletrônico) Paratrichodina africana (Ciliophora: Trichodinidae) of wild and cultured Nile tilapia in the Northern Brazil Paratrichodina africana (Ciliophora: Trichodinidae) de tilápia do Nilo selvagem e cultivada no Norte do Brasil Marcos Tavares-Dias1; Natália da Costa Marchiori2; Maurício Laterça Martins2* 1Laboratório de Aquicultura e Pesca, Embrapa Amapá, Macapá, AP, Brasil 2Laboratório de Sanidade em Organismos Aquáticos – AQUOS, Departamento de Aquicultura, Universidade Federal de Santa Catarina – UFSC, Florianópolis, SC, Brasil Received September 8, 2012 Accepted February 1, 2013 Abstract The present work morphologically characterizes Paratrichodina africana from the gills of wild and farmed Nile tilapia from Northern Brazil (eastern Amazonia). Ninety fish were captured for parasitological analysis in Macapá, State of Amapá, from a wetland area bathed by the Amazon River commonly called ‘Ressaca do Zerão’ (n = 52), as well as from a local fish farm (n = 38). -

Handbook of Shrimp Diseases
LOAN COPY ONLY TAMU-H-95-001 C3 Handbook of Shrimp Diseases Aquaculture S.K. Johnson Department of Wildlife and Fisheries Sciences Texas A&M University 90-601 (rev) Introduction 2 Shrimp Species 2 Shrimp Anatomy 2 Obvious Manifestations ofShrimp Disease 3 Damaged Shells , 3 Inflammation and Melanization 3 Emaciation and Nutritional Deficiency 4 Muscle Necrosis 5 Tumors and Other Tissue Problems 5 Surface Fouling 6 Cramped Shrimp 6 Unusual Behavior 6 Developmental Problems 6 Growth Problems 7 Color Anomalies 7 Microbes 8 Viruses 8 Baceteria and Rickettsia 10 Fungus 12 Protozoa 12 Haplospora 13 Gregarina 15 Body Invaders 16 Surface Infestations 16 Worms 18 Trematodes 18 Cestodes 18 Nematodes 18 Environment 20 Publication of this handbook is a coop erative effort of the Texas A&M Univer sity Sea Grant College Program, the Texas A&M Department of Wildlife and $2.00 Fisheries Sciences and the Texas Additional copies available from: Agricultural Extension Service. Produc Sea Grant College Program tion is supported in part by Institutional 1716 Briarcrest Suite 603 Grant No. NA16RG0457-01 to Texas Bryan, Texas 77802 A&M University by the National Sea TAMU-SG-90-601(r) Grant Program, National Oceanic and 2M August 1995 Atmospheric Administration, U.S. De NA89AA-D-SG139 partment of Commerce. A/1-1 Handbook ofShrimp Diseases S.K. Johnson Extension Fish Disease Specialist This handbook is designed as an information source and tail end (abdomen). The parts listed below are apparent upon field guide for shrimp culturists, commercial fishermen, and outside examination (Fig. 1). others interested in diseases or abnormal conditions of shrimp.