NIK Controls Classical and Alternative NF-Kb Activation and Is Necessary for the Survival of Human T-Cell Lymphoma Cells
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Epigenetic Reprogramming Sensitizes CML Stem Cells to Combined EZH2 and Tyrosine
Published OnlineFirst September 14, 2016; DOI: 10.1158/2159-8290.CD-16-0263 RESEARCH BRIEF Epigenetic Reprogramming Sensitizes CML Stem Cells to Combined EZH2 and Tyrosine Kinase Inhibition Mary T. Scott 1 , 2 , Koorosh Korfi 1 , 2 , Peter Saffrey 1 , Lisa E.M. Hopcroft 2 , Ross Kinstrie 1 , Francesca Pellicano 2 , Carla Guenther 1 , 2 , Paolo Gallipoli 2 , Michelle Cruz 1 , Karen Dunn 2 , Heather G. Jorgensen 2 , Jennifer E. Cassels 2 , Ashley Hamilton 2 , Andrew Crossan 1 , Amy Sinclair 2 , Tessa L. Holyoake 2 , and David Vetrie 1 ABSTRACT A major obstacle to curing chronic myeloid leukemia (CML) is residual disease main- tained by tyrosine kinase inhibitor (TKI)–persistent leukemic stem cells (LSC). These are BCR–ABL1 kinase independent, refractory to apoptosis, and serve as a reservoir to drive relapse or TKI resistance. We demonstrate that Polycomb Repressive Complex 2 is misregulated in chronic phase CML LSCs. This is associated with extensive reprogramming of H3K27me3 targets in LSCs, thus sensi- tizing them to apoptosis upon treatment with an EZH2-specifi c inhibitor (EZH2i). EZH2i does not impair normal hematopoietic stem cell survival. Strikingly, treatment of primary CML cells with either EZH2i or TKI alone caused signifi cant upregulation of H3K27me3 targets, and combined treatment further potentiated these effects and resulted in signifi cant loss of LSCs compared to TKI alone, in vitro , and in long-term bone marrow murine xenografts. Our fi ndings point to a promising epigenetic-based thera- peutic strategy to more effectively target LSCs in patients with CML receiving TKIs. SIGNIFICANCE: In CML, TKI-persistent LSCs remain an obstacle to cure, and approaches to eradicate them remain a signifi cant unmet clinical need. -

Gene Essentiality Landscape and Druggable Oncogenic Dependencies in Herpesviral Primary Effusion Lymphoma
ARTICLE DOI: 10.1038/s41467-018-05506-9 OPEN Gene essentiality landscape and druggable oncogenic dependencies in herpesviral primary effusion lymphoma Mark Manzano1, Ajinkya Patil1, Alexander Waldrop2, Sandeep S. Dave2, Amir Behdad3 & Eva Gottwein1 Primary effusion lymphoma (PEL) is caused by Kaposi’s sarcoma-associated herpesvirus. Our understanding of PEL is poor and therefore treatment strategies are lacking. To address this 1234567890():,; need, we conducted genome-wide CRISPR/Cas9 knockout screens in eight PEL cell lines. Integration with data from unrelated cancers identifies 210 genes as PEL-specific oncogenic dependencies. Genetic requirements of PEL cell lines are largely independent of Epstein-Barr virus co-infection. Genes of the NF-κB pathway are individually non-essential. Instead, we demonstrate requirements for IRF4 and MDM2. PEL cell lines depend on cellular cyclin D2 and c-FLIP despite expression of viral homologs. Moreover, PEL cell lines are addicted to high levels of MCL1 expression, which are also evident in PEL tumors. Strong dependencies on cyclin D2 and MCL1 render PEL cell lines highly sensitive to palbociclib and S63845. In summary, this work comprehensively identifies genetic dependencies in PEL cell lines and identifies novel strategies for therapeutic intervention. 1 Department of Microbiology-Immunology, Feinberg School of Medicine, Northwestern University, Chicago, IL 60611, USA. 2 Duke Cancer Institute and Center for Genomic and Computational Biology, Duke University, Durham, NC 27708, USA. 3 Department of Pathology, Feinberg School of Medicine, Northwestern University, Chicago, IL 60611, USA. Correspondence and requests for materials should be addressed to E.G. (email: [email protected]) NATURE COMMUNICATIONS | (2018) 9:3263 | DOI: 10.1038/s41467-018-05506-9 | www.nature.com/naturecommunications 1 ARTICLE NATURE COMMUNICATIONS | DOI: 10.1038/s41467-018-05506-9 he human oncogenic γ-herpesvirus Kaposi’s sarcoma- (IRF4), a critical oncogene in multiple myeloma33. -

Supporting Online Material
1 2 3 4 5 6 7 Supplementary Information for 8 9 Fractalkine-induced microglial vasoregulation occurs within the retina and is altered early in diabetic 10 retinopathy 11 12 *Samuel A. Mills, *Andrew I. Jobling, *Michael A. Dixon, Bang V. Bui, Kirstan A. Vessey, Joanna A. Phipps, 13 Ursula Greferath, Gene Venables, Vickie H.Y. Wong, Connie H.Y. Wong, Zheng He, Flora Hui, James C. 14 Young, Josh Tonc, Elena Ivanova, Botir T. Sagdullaev, Erica L. Fletcher 15 * Joint first authors 16 17 Corresponding author: 18 Prof. Erica L. Fletcher. Department of Anatomy & Neuroscience. The University of Melbourne, Grattan St, 19 Parkville 3010, Victoria, Australia. 20 Email: [email protected] ; Tel: +61-3-8344-3218; Fax: +61-3-9347-5219 21 22 This PDF file includes: 23 24 Supplementary text 25 Figures S1 to S10 26 Tables S1 to S7 27 Legends for Movies S1 to S2 28 SI References 29 30 Other supplementary materials for this manuscript include the following: 31 32 Movies S1 to S2 33 34 35 36 1 1 Supplementary Information Text 2 Materials and Methods 3 Microglial process movement on retinal vessels 4 Dark agouti rats were anaesthetized, injected intraperitoneally with rhodamine B (Sigma-Aldrich) to label blood 5 vessels and retinal explants established as described in the main text. Retinal microglia were labelled with Iba-1 6 and imaging performed on an inverted confocal microscope (Leica SP5). Baseline images were taken for 10 7 minutes, followed by the addition of PBS (10 minutes) and then either fractalkine or fractalkine + candesartan 8 (10 minutes) using concentrations outlined in the main text. -
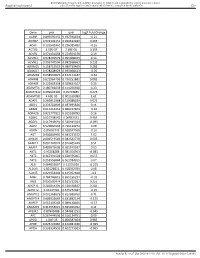
Gene Pval Qval Log2 Fold Change AAMP 0.895690332 0.952598834
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) Gut Gene pval qval Log2 Fold Change AAMP 0.895690332 0.952598834 -0.21 ABI3BP 0.002302151 0.020612283 0.465 ACHE 0.103542461 0.296385483 -0.16 ACTG2 2.99E-07 7.68E-05 3.195 ACVR1 0.071431098 0.224504378 0.19 ACVR1C 0.978209579 0.995008423 0.14 ACVRL1 0.006747504 0.042938663 0.235 ADAM15 0.158715519 0.380719469 0.285 ADAM17 0.978208929 0.995008423 -0.05 ADAM28 0.038932876 0.152174187 -0.62 ADAM8 0.622964796 0.790251882 0.085 ADAM9 0.122003358 0.329623107 0.25 ADAMTS1 0.180766659 0.414256926 0.23 ADAMTS12 0.009902195 0.05703885 0.425 ADAMTS8 4.60E-05 0.001169089 1.61 ADAP1 0.269811968 0.519388039 0.075 ADD1 0.233702809 0.487695826 0.11 ADM2 0.012213453 0.066227879 -0.36 ADRA2B 0.822777921 0.915518785 0.16 AEBP1 0.010738542 0.06035531 0.465 AGGF1 0.117946691 0.320915024 -0.095 AGR2 0.529860903 0.736120272 0.08 AGRN 0.85693743 0.928047568 -0.16 AGT 0.006849995 0.043233572 1.02 AHNAK 0.006519543 0.042542779 0.605 AKAP12 0.001747074 0.016405449 0.51 AKAP2 0.409929603 0.665919397 0.05 AKT1 0.95208288 0.985354963 -0.085 AKT2 0.367391504 0.620376005 0.055 AKT3 0.253556844 0.501934205 0.07 ALB 0.064833867 0.21195036 -0.315 ALDOA 0.83128831 0.918352939 0.08 ALOX5 0.029954404 0.125352668 -0.3 AMH 0.784746815 0.895196237 -0.03 ANG 0.050500474 0.181732067 0.255 ANGPT1 0.281853305 0.538528647 0.285 ANGPT2 0.43147281 0.675272487 -0.15 ANGPTL2 0.001368876 0.013688762 0.71 ANGPTL4 0.686032669 0.831882134 -0.175 ANPEP 0.019103243 0.089148466 -0.57 ANXA2P2 0.412553021 0.665966092 0.11 AP1M2 0.87843088 0.944681253 -0.045 APC 0.267444505 0.516134751 0.09 APOD 1.04E-05 0.000587404 0.985 APOE 0.023722987 0.104981036 -0.395 APOH 0.336334555 0.602273505 -0.065 Sundar R, et al. -

R&D Assay for Alzheimer's Disease
R&DR&D assayassay forfor Alzheimer’sAlzheimer’s diseasedisease Target screening⳼ Ⲽ㬔 antibody array, ᢜ⭉㬔 ⸽ἐⴐ Amyloid β-peptide Alzheimer’s disease⯸ ኸᷠ᧔ ᆹ⸽ inhibitor, antibody, ELISA kit Surwhrph#Surilohu#Dqwlerg|#Duud| 6OUSFBUFE 1."5SFBUFE )41 $3&# &3, &3, )41 $3&# &3, &3, 壤伡庰䋸TBNQMF ɅH 侴䋸嵄䍴䋸BOBMZUFT䋸䬱娴哜塵 1$ 1$ 1$ 1$ 5IFNPTUSFGFSFODFEBSSBZT 1$ 1$ QQ α 34, .4, 503 Q α 34, .4, 503 %SVHTDSFFOJOH0òUBSHFUFòFDUT0ATHWAY涭廐 6OUSFBUFE 堄币䋸4BNQMF侴䋸8FTUFSOPS&-*4"䍘䧽 1."5SFBUFE P 8FTUFSOCMPU廽喜儤应侴䋸0, Z 4VCTUSBUF -JHIU )31DPOKVHBUFE1BO "OUJQIPTQIPUZSPTJOF .FBO1JYFM%FOTJUZ Y $BQUVSF"OUJCPEZ 5BSHFU"OBMZUF "SSBZ.FNCSBOF $3&# &3, &3, )41 .4, Q α 34, 503 Human XL Cytokine Array kit (ARY022, 102 analytes) Adiponectin,Aggrecan,Angiogenin,Angiopoietin-1,Angiopoietin-2,BAFF,BDNF,Complement,Component C5/C5a,CD14,CD30,CD40L, Chitinase 3-like 1,Complement Factor D,C-Reactive Protein,Cripto-1,Cystatin C,Dkk-1,DPPIV,EGF,EMMPRIN,ENA-78,Endoglin, Fas L,FGF basic,FGF- 7,FGF-19,Flt-3 L,G-CSF,GDF-15,GM-CSF,GRO-α,Grow th Hormone,HGF,ICAM-1,IFN-γ,IGFBP-2,IGFBP-3, IL-1α,IL-1β, IL-1ra,IL-2,IL-3,IL-4,IL- 5,IL-6,IL-8, IL-10,IL-11,IL-12, IL-13,IL-15,IL-16,IL-17A,IL-18 BPa,IL-19,IL-22, IL-23,IL-24,IL-27, IL-31,IL-32α/β/γ,IL-33,IL-34,IP-10,I-TAC,Kallikrein 3,Leptin,LIF,Lipocalin-2,MCP-1,MCP-3,M-CSF,MIF,MIG,MIP-1α/MIP-1β,MIP-3α,MIP-3β,MMP-9, Myeloperoxidase,Osteopontin, p70, PDGF-AA, PDGF-AB/BB,Pentraxin-3, PF4, RAGE, RANTES,RBP4,Relaxin-2, Resistin,SDF-1α,Serpin E1, SHBG, ST2, TARC,TFF3,TfR,TGF- ,Thrombospondin-1,TNF-α, uPAR, VEGF, Vitamin D BP Human Protease (34 analytes) / -

C-Terminal Region (ARP58987 P050) Data Sheet
CASP3 antibody - C-terminal region (ARP58987_P050) Data Sheet Product Number ARP58987_P050 Product Name CASP3 antibody - C-terminal region (ARP58987_P050) Size 50ug Gene Symbol CASP3 Alias Symbols CPP32; CPP32B; SCA-1 Nucleotide Accession# NM_032991 Protein Size (# AA) 277 amino acids Molecular Weight 12kDa Product Format Lyophilized powder NCBI Gene Id 836 Host Rabbit Clonality Polyclonal Official Gene Full Name Caspase 3, apoptosis-related cysteine peptidase Gene Family CASP This is a rabbit polyclonal antibody against CASP3. It was validated on Western Blot by Aviva Systems Biology. At Aviva Systems Biology we manufacture rabbit polyclonal antibodies on a large scale (200-1000 Description products/month) of high throughput manner. Our antibodies are peptide based and protein family oriented. We usually provide antibodies covering each member of a whole protein family of your interest. We also use our best efforts to provide you antibodies recognize various epitopes of a target protein. For availability of antibody needed for your experiment, please inquire (). Peptide Sequence Synthetic peptide located within the following region: NLKYEVRNKNDLTREEIVELMRDVSKEDHSKRSSFVCVLLSHGEEGIIFG CASP3 is a protein which is a member of the cysteine-aspartic acid protease (caspase) family. Sequential activation of caspases plays a central role in the execution-phase of cell apoptosis. Caspases exist as inactive proenzymes which undergo proteolytic processing at conserved aspartic residues to produce two subunits, Description of Target large and small, that dimerize to form the active enzyme. This protein cleaves and activates caspases 6, 7 and 9, and the protein itself is processed by caspases 8, 9 and 10. It is the predominant caspase involved in the cleavage of amyloid-beta 4A precursor protein, which is associated with neuronal death in Alzheimer's disease. -

Supplementary Material DNA Methylation in Inflammatory Pathways Modifies the Association Between BMI and Adult-Onset Non- Atopic
Supplementary Material DNA Methylation in Inflammatory Pathways Modifies the Association between BMI and Adult-Onset Non- Atopic Asthma Ayoung Jeong 1,2, Medea Imboden 1,2, Akram Ghantous 3, Alexei Novoloaca 3, Anne-Elie Carsin 4,5,6, Manolis Kogevinas 4,5,6, Christian Schindler 1,2, Gianfranco Lovison 7, Zdenko Herceg 3, Cyrille Cuenin 3, Roel Vermeulen 8, Deborah Jarvis 9, André F. S. Amaral 9, Florian Kronenberg 10, Paolo Vineis 11,12 and Nicole Probst-Hensch 1,2,* 1 Swiss Tropical and Public Health Institute, 4051 Basel, Switzerland; [email protected] (A.J.); [email protected] (M.I.); [email protected] (C.S.) 2 Department of Public Health, University of Basel, 4001 Basel, Switzerland 3 International Agency for Research on Cancer, 69372 Lyon, France; [email protected] (A.G.); [email protected] (A.N.); [email protected] (Z.H.); [email protected] (C.C.) 4 ISGlobal, Barcelona Institute for Global Health, 08003 Barcelona, Spain; [email protected] (A.-E.C.); [email protected] (M.K.) 5 Universitat Pompeu Fabra (UPF), 08002 Barcelona, Spain 6 CIBER Epidemiología y Salud Pública (CIBERESP), 08005 Barcelona, Spain 7 Department of Economics, Business and Statistics, University of Palermo, 90128 Palermo, Italy; [email protected] 8 Environmental Epidemiology Division, Utrecht University, Institute for Risk Assessment Sciences, 3584CM Utrecht, Netherlands; [email protected] 9 Population Health and Occupational Disease, National Heart and Lung Institute, Imperial College, SW3 6LR London, UK; [email protected] (D.J.); [email protected] (A.F.S.A.) 10 Division of Genetic Epidemiology, Medical University of Innsbruck, 6020 Innsbruck, Austria; [email protected] 11 MRC-PHE Centre for Environment and Health, School of Public Health, Imperial College London, W2 1PG London, UK; [email protected] 12 Italian Institute for Genomic Medicine (IIGM), 10126 Turin, Italy * Correspondence: [email protected]; Tel.: +41-61-284-8378 Int. -

Supplementary Material and Methods
Supplementary material and methods Generation of cultured human epidermal sheets Normal human epidermal keratinocytes were isolated from human breast skin. Keratinocytes were grown on a feeder layer of irradiated human fibroblasts pre-seeded at 4000 cells /cm² in keratinocyte culture medium (KCM) containing a mix of 3:1 DMEM and HAM’s F12 (Invitrogen, Carlsbad, USA), supplemented with 10% FCS, 10ng/ml epidermal growth factor (EGF; R&D systems, Minneapolis, MN, USA), 0.12 IU/ml insulin (Lilly, Saint- Cloud, France), 0.4 mg/ml hydrocortisone (UpJohn, St Quentin en Yvelelines, France) , 5 mg/ml triiodo-L- thyronine (Sigma, St Quentin Fallavier, France), 24.3 mg/ml adenine (Sigma), isoproterenol (Isuprel, Hospira France, Meudon, France) and antibiotics (20 mg/ml gentamicin (Phanpharma, Fougères, France), 100 IU/ml penicillin (Phanpharma), and 1 mg/ml amphotericin B (Phanpharma)). The medium was changed every two days. NHEK were then cultured over a period of 13 days according to the protocol currently used at the Bank of Tissues and Cells for the generation of clinical grade epidermal sheets used for the treatment of severe extended burns (Ref). When needed, cells were harvested with trypsin-EDTA 0.05% (Thermo Fisher Scientific, Waltham, MA, USA) and collected for analysis. Clonogenic assay Keratinocytes were seeded on a feeder layer of irradiated fibroblasts, at a clonal density of 10-20 cells/cm² and cultivated for 10 to 14 days. Three flasks per tested condition were fixed and colored in a single 30 mns step using rhodamine B (Sigma) diluted at 0.01 g/ml in 4% paraformaldehyde. In each tested condition, cells from 3 other flasks were numerated after detachment by trypsin treatment. -

The Human Cytomegalovirus–Encoded Chemokine Receptor US28 Promotes Angiogenesis and Tumor Formation Via Cyclooxygenase-2
Published OnlineFirst March 24, 2009; DOI: 10.1158/0008-5472.CAN-08-2487 Research Article The Human Cytomegalovirus–Encoded Chemokine Receptor US28 Promotes Angiogenesis and Tumor Formation via Cyclooxygenase-2 David Maussang,1 Ellen Langemeijer,1 Carlos P. Fitzsimons,1 Marijke Stigter-van Walsum,2 Remco Dijkman,3 Martin K. Borg,3 Erik Slinger,1 Andreas Schreiber,4 Detlef Michel,4 Cornelis P. Tensen,3 Guus A.M.S. van Dongen,2 Rob Leurs,1 and Martine J. Smit1 1Leiden/Amsterdam Center for Drug Research, Division of Medicinal Chemistry, Faculty of Sciences, Vrije Universiteit Amsterdam; 2Department of Otolaryngology/Head and Neck Surgery, Vrije Universiteit Medical Center, Amsterdam, the Netherlands; 3Department of Dermatology, Leiden University Medical Center, Leiden, the Netherlands; and 4Institute of Virology, University Clinic Ulm, Ulm, Germany Abstract considered an oncogenic herpesvirus (5), but it has been suggested The human cytomegalovirus (HCMV), potentially associated to act as an oncomodulator (6). The presence of HCMV proteins with the development of malignancies, encodes the constitu- has been detected in several malignancies, such as colon cancer (7), tively active chemokine receptor US28. Previously, we have malignant glioblastoma (8), and breast cancer (9). Importantly, shown that US28 expression induces an oncogenic phenotype HCMV preferably infects tumor cells, leading to enhanced cell both in vitro and in vivo. Microarray analysis revealed proliferation, angiogenesis, and resistance to apoptosis (6, 10). differential expression of genes involved in oncogenic signal- Of interest, most herpesviruses contain one or more genes that ing in US28-expressing NIH-3T3 cells. In particular, the encode for constitutively active G-protein coupled receptors (1). -

ADAM8 Expression in Invasive Breast Cancer Promotes Tumor Dissemination and Metastasis
ADAM8 expression in invasive breast cancer promotes tumor dissemination and metastasis Mathilde Romagnoli, Nora Mineva, Michael Polmear, Catharina Conrad, Srimathi Srinivasan, Delphine Loussouarn, Sophie Barillé-Nion, Irene Georgakoudi, Aine Dagg, Enda Mcdermott, et al. To cite this version: Mathilde Romagnoli, Nora Mineva, Michael Polmear, Catharina Conrad, Srimathi Srinivasan, et al.. ADAM8 expression in invasive breast cancer promotes tumor dissemination and metastasis. EMBO Molecular Medicine, Wiley Open Access, 2014, 6 (2), pp.278-294. 10.1002/emmm.201303373. inserm-02447040 HAL Id: inserm-02447040 https://www.hal.inserm.fr/inserm-02447040 Submitted on 21 Jan 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Research Article ADAM8 expression in invasive breast cancer promotes tumor dissemination and metastasis Mathilde Romagnoli1, Nora D Mineva1, Michael Polmear2, Catharina Conrad3, Srimathi Srinivasan1, Delphine Loussouarn4, Sophie Barille-Nion4, Irene Georgakoudi2, Aine Dagg5,6, Enda W McDermott6, Michael J Duffy6, Patricia M. McGowan5,6, Uwe Schlomann3, Maddy Parsons7,Jorg€ W Bartsch3 & Gail E Sonenshein1,* Abstract Introduction The transmembrane metalloprotease-disintegrin ADAM8 mediates Cancer metastasis results from a multistep process that selects for cell adhesion and shedding of ligands, receptors and extracellular invasive tumor cells capable of escaping from the primary site and matrix components. -

Airway Inflammation in Mice Adam8 Limits the Development of Allergic
Adam8 Limits the Development of Allergic Airway Inflammation in Mice Martin D. Knolle, Takahiro Nakajima, Anja Hergrueter, Kushagra Gupta, Francesca Polverino, Vanessa J. Craig, This information is current as Susanne E. Fyfe, Muhammad Zahid, Perdita Permaul, of October 2, 2021. Manuela Cernadas, Gilbert Montano, Yohannes Tesfaigzi, Lynette Sholl, Lester Kobzik, Elliot Israel and Caroline A. Owen J Immunol 2013; 190:6434-6449; Prepublished online 13 May 2013; Downloaded from doi: 10.4049/jimmunol.1202329 http://www.jimmunol.org/content/190/12/6434 http://www.jimmunol.org/ Supplementary http://www.jimmunol.org/content/suppl/2013/05/13/jimmunol.120232 Material 9.DC1 References This article cites 77 articles, 18 of which you can access for free at: http://www.jimmunol.org/content/190/12/6434.full#ref-list-1 Why The JI? Submit online. by guest on October 2, 2021 • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology Adam8 Limits the Development of Allergic Airway Inflammation in Mice Martin D. -

MIND Bomb 2 Prevents RIPK1 Kinase Activity-Dependent and -Independent Apoptosis Through Ubiquitylation of Cflipl
ARTICLE https://doi.org/10.1038/s42003-020-01603-y OPEN MIND bomb 2 prevents RIPK1 kinase activity-dependent and -independent apoptosis through ubiquitylation of cFLIPL Osamu Nakabayashi 1, Hirotaka Takahashi2, Kenta Moriwaki1, Sachiko Komazawa-Sakon1, Fumiaki Ohtake3, 1234567890():,; Shin Murai 1, Yuichi Tsuchiya1, Yuki Koyahara1, Yasushi Saeki 4, Yukiko Yoshida4, Soh Yamazaki1, ✉ Fuminori Tokunaga 5, Tatsuya Sawasaki 2 & Hiroyasu Nakano 1 Mind bomb 2 (MIB2) is an E3 ligase involved in Notch signalling and attenuates TNF-induced apoptosis through ubiquitylation of receptor-interacting protein kinase 1 (RIPK1) and cylin- dromatosis. Here we show that MIB2 bound and conjugated K48– and K63–linked poly- ubiquitin chains to a long-form of cellular FLICE-inhibitory protein (cFLIPL), a catalytically inactive homologue of caspase 8. Deletion of MIB2 did not impair the TNF-induced complex I formation that mediates NF-κB activation but significantly enhanced formation of cytosolic death-inducing signalling complex II. TNF-induced RIPK1 Ser166 phosphorylation, a hallmark of RIPK1 death-inducing activity, was enhanced in MIB2 knockout cells, as was RIPK1 kinase activity-dependent and -independent apoptosis. Moreover, RIPK1 kinase activity-independent apoptosis was induced in cells expressing cFLIPL mutants lacking MIB2-dependent ubiqui- tylation. Together, these results suggest that MIB2 suppresses both RIPK1 kinase activity- dependent and -independent apoptosis, through suppression of RIPK1 kinase activity and ubiquitylation of cFLIPL, respectively. 1 Department of Biochemistry, Toho University School of Medicine, 5-21-16 Omori-Nishi, Ota-ku, Tokyo 143-8540, Japan. 2 Division of Cell-Free Sciences, Proteo-Science Center (PROS), 3 Bunkyo-cho, Matsuyama, Ehime 790-8577, Japan.