Servicios GENYCA 20171218 ENGLISH
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

DIAGMOL Town, ZIPCODE :…………………………………………………… Director : Prof
Mr. Mrs. (IN UPPER CASE, please) Name:……………………………………………………. Maiden Name:……………………………………. DMGL / Service de Médecine Génétique Centre d’accueil des prélèvements (CAP) First name :………………………………………………… Bâtiment des Laboratoires (BATLab), local 8D-0-850.1 Date of birth : ............ / ........... / …………… 4 rue Gabrielle-Perret-Gentil, 1211 Genève 14 Legal representative (for minors) : father mother Molecular and Genomic Diagnostics Laboratory Name/first name :…………………………………………… Street/N°:……………………………………………………… http://www.hug-ge.ch/feuilles-de-demande DIAGMOL Town, ZIPCODE :…………………………………………………… Director : Prof. Stylianos ANTONARAKIS Hospitalisation Unit: …………… Physician :…………………… Lab managers: N° EdS : …………………………………………………………… Dr J.-L. BLOUIN, Dr Th. NOUSPIKEL, Dr. M. GUIPPONI Invoice address: Patient Prescriptor Insurance [email protected], [email protected], [email protected] Type of case : Disease AI Accident Pregnancy Lab direct or results: Phone/FAX: +41 (0) 22 37 21 826 / 21 860 N° AVS (Mandatory for AI) : ………………………...................... Sample Entrance Center (CAP) : Phone +41 (0) 22 37 21 800 Insurance : ………………… Insured N° : ……………………… PHYSICIAN PHYSICIAN (NAME/First name - Street/N°- Town, ZIPCODE - Phone/FAX. IN UPPER CASES, PLEASE) COPY TO OTHER PHYSICIAN (NAME/First name - Street/N°- Town, ZIPCODE - Phone/FAX. IN UPPER CASES, PLEASE) « The laboratory is granted permission by the Physician/Patient to transmit copies of the report to other physicians» Opposition of the patient to the registration of this request results in the electronic patient record (DPI) of the HUG If the patient belongs to a family already known to the laboratory, please indicate index case NAME: CLINICAL INFORMATIONS given by the physician: Ethnic origins Father Mother Currently pregnant Date of last menses Number of weeks of amenorrhea SAMPLE(S) Most of our tests work from 4 ml of blood in EDTA (children <2 ans : 1 On every and single NAME First name ml : ok) or from purified DNA (some exceptions apply for some tests) tube. -

Jeffrey Kleinberger Ph.D. Candidate Molecular Medicine Program, Genome Biology Track Graduate Program in Life Sciences, University of Maryland, Baltimore
Discovery and Analysis of Patients with Monogenic Diabetes in Multiple Cohorts to Guide Future Diagnosis Item Type dissertation Authors Kleinberger, Jeffrey Publication Date 2017 Abstract Monogenic diabetes is hyperglycemia caused by a variant in a single gene, and it accounts for approximately 1-2% of all diabetes cases. A genetic diagnosis of monogenic diabetes is important because the most common gene etiologies can be effectively ... Keywords MODY; monogenic diabetes; screening; Clinical Trial; Diabetes Mellitus, Type 2; Zebrafish Download date 05/10/2021 09:00:42 Link to Item http://hdl.handle.net/10713/7072 Jeffrey Kleinberger Ph.D. Candidate Molecular Medicine Program, Genome Biology Track Graduate Program in Life Sciences, University of Maryland, Baltimore Contact Information Medical School Teaching Facility 357 685 West Baltimore St. Baltimore, MD 21201 [email protected] Education Juniata College Huntingdon, PA August 2004 – May 2008 Cum Laude distinction (GPA: 3.701) Biochemistry major Coursework included: Organic Chemistry, Bioinorganic Chemistry, Analytical Chemistry, Physical Chemistry, Chemistry Research, Biology, Cell Biology, Genetics, Biostatistics, Molecular Techniques, Physiology, Microbiology, Biochemistry and Molecular Biology (I-III), Calculus (I&II), Physics (I&II), Scientific Glassblowing, College Writing Seminar, Art of Public Speaking, Intro to Psychology, Learning & Conditioning, Mod. Knowledge & The Self, United States to 1877, The American Revolution, Survey of Western Art, Modern Architecture, -

一般演題(口演)/Oral Sessions
一般演題(口演)!Oral Sessions ご ! 案 第 60 回大会賞候補セッション(口演) Oral Presentation Award Session 内 日 時:10 月 16 日(金)11:25~12:20 第 2 会場(南館 4 階 錦) 座 長:稲澤 譲治(東京医科歯科大学難治疾患研究所ゲノム応用医学研究部門(分子細胞遺伝)) 田中 敏博(東京医科歯科大学疾患バイオリソースセンター) Date:Oct. 16 (Fri.) 11:25~12:20 Room 2(Nishiki, South Tower 4F) Chairs:Johji Inazawa(Tokyo Medical and Dental University) Toshihiro Tanaka(Tokyo Medical and Dental University) プ ロ BO-1 ゲノムワイド関連解析(GWAS)データを活用した in silico 解析による新規抗パーキンソン病薬 グ の探索 ラ In silico drug discovery for Parkinson’s disease by using genome!wide association study ム (GWAS)data ○上中 健 1(Takeshi Uenaka)、佐竹 渉 1(Wataru Satake)、謝 珮琴 1(Pei"Chieng Cha)、 岡田 随象 2(Yukinori Okada)、戸田 達史 1(Tatsushi Toda) 1 神戸大学大学院医学研究科神経内科!分子脳科学 指 (Division of Neurology, Kobe University Graduate School of Medicine, Kobe, Japan) 定 2 東京医科歯科大学大学院医歯学総合研究科疾患多様性遺伝学分野 演 (Department of Human Genetics and Disease Diversity, Graduate School of Medical and Dental Sciences, 題 Tokyo Medical and Dental University, Tokyo, Japan) 抄 録 BO-2 MIRAGE 症候群:機能亢進型 SAMD9 変異を原因とする新規症候群の発見 MIRAGE syndrome:New genetic syndrome caused by activating SAMD9 mutations ○鳴海 覚志 1(Satoshi Narumi)、天野 直子 1(Naoko Amano)、石井 智弘 1(Tomohiro Ishii)、 一 勝又 規行 2(Noriyuki Katsumata)、福澤 龍二 3(Ryuji Fukuzawa)、芝田 晋介 4(Shinsuke Shibata)、 般 4 5 6 演 岡野 栄之 (Hideyuki Okano)、清水 厚志 (Atsushi Shimizu)、三宅 紀子 (Noriko Miyake)、 題 松本 直通 6(Naomichi Matsumoto)、長谷川 奉延 1(Tomonobu Hasegawa) ( 1 慶應義塾大学医学部小児科 口 (Department of Pediatrics, Keio University School of Medicine, Tokyo, Japan) 演 ) 2 国立成育医療研究センター研究所分子内分泌研究部 抄 (Department of Molecular Endocrinology, National Research -

UNSCEAR 2001 Report to the General Assembly, with Scientific Annex
HEREDITARY EFFECTS OF RADIATION United Nations Scientific Committee on the Effects of Atomic Radiation UNSCEAR 2001 Report to the General Assembly, with Scientific Annex UNITED NATIONS HEREDITARY EFFECTS OF RADIATION United Nations Scientific Committee on the Effects of Atomic Radiation UNSCEAR 2001 Report to the General Assembly, with Scientific Annex UNITED NATIONS New York, 2001 NOTE The report of the Committee without its scientific annex appears as Official Records of the General Assembly. Fifty-sixth Session, Supplement No. 46 (N56146). The designation employed and the presentation of material in this publication do not imply the expression of any opinion whatsoever on the pad of the Secretariat of the United Nationsconcerning the legal status of any country, temtory, city orarea, or of its authorities, or concerning the delimitation of its frontiers or boundaries. The country names used in this document are, In most cases, those that were in use at the time the data were collected or the text prepared. In othercases, however, the names have been updated, where this was posslble and appropriate, to reflect political changes. UNITED NATIONS PUBLICATION Sales No. E.O1 .IX.2 ISBN 92-1-1 42244-2 Report of the United Nations Scientific Committee on the Effects of Atomic Radiation to the General Assembly 1. During the past few years, the United Nations 4. The Committee wishes to acknowledge the assistance Scientific Committee on the Effects of Atomic Radiation1 of the consultant, K. Sankaranarayanan, in the preparation has undertaken broad reviews of the sources and effects of of the scientific annex and the advice of the international ionizing radiation. -

Uncommon Forms of Diabetes
Clinical Medicine 2021 Vol 21, No 4: e337–41 CME: DIABETES Uncommon forms of diabetes Authors: Yun-Ni LeeA and Mohammed SB HudaB Diabetes mellitus is a common condition which all clinicians and insulin independence. It is estimated to account for 1%–2% will encounter in their clinical practice. The most common of patients diagnosed with diabetes and, in the UK, the prevalence form is type 2 diabetes followed by type 1 diabetes. However, of MODY is estimated to be at 108 cases per million.3 However, there are many other atypical forms of diabetes which are it may be a significant underestimate and these figures are not important for a clinician to consider as it can impact on the accurate until large population screening studies are performed. ABSTRACT diagnosis and their management. The most common mutations are hepatocyte nuclear factor-1- This article focuses on maturity onset diabetes of the young alpha (HNF1α; 52%), glucokinase (GCK; 32%) and HNF4α (10%), (MODY), latent autoimmune diabetes in adults (LADA), see Table 2.3 ketosis-prone diabetes and other secondary forms of diabetes such as pancreatic cancer and haemochromatosis. We briefly Hepatocyte nuclear factor-1-alpha gene describe the key clinical features of these forms of diabetes and their investigations and treatment. Formerly called MODY3, mutations on the HNF1α gene on chromosome 3 are associated with a progressive defect of insulin secretion.4 Mutations here also result in low renal threshold for 5 Introduction glucose and thus mutation carriers have detectable glycosuria. In the UK, around 90% of people with diabetes have type 2 diabetes (T2D), around 8% have type 1 diabetes (T1D) and around 2% have other forms of diabetes.1 Key points Typically, we see T1D present in a young, lean patient with Suspect other uncommon forms of diabetes if the clinical marked symptoms of polyuria, polydipsia, weight loss and diabetic picture does not fit type 1 or type 2 diabetes. -

Mat Kadi Tora Tutti O Al Ut Hit Hitta Atuh
MAT KADI TORA TUTTI USO AL20180235194A1 UT HIT HITTA ATUH ( 19) United States (12 ) Patent Application Publication ( 10) Pub . No. : US 2018 /0235194 A1 Fahrenkrug et al. (43 ) Pub . Date : Aug . 23, 2018 ( 54 ) MULTIPLEX GENE EDITING Publication Classification (51 ) Int. Ci. ( 71 ) Applicant: Recombinetics , Inc ., Saint Paul, MN A01K 67/ 027 (2006 . 01 ) (US ) C12N 15 / 90 ( 2006 .01 ) (72 ) Inventors : Scott C . Fahrenkrug, Minneapolis , (52 ) U . S . CI. MN (US ) ; Daniel F . Carlson , CPC .. .. A01K 67 / 0276 (2013 . 01 ) ; C12N 15 / 907 Woodbury , MN (US ) ( 2013 .01 ) ; A01K 67 /0275 ( 2013 .01 ) ; A01K 2267/ 02 (2013 .01 ) ; AOIK 2217 / 15 (2013 .01 ) ; AOIK 2227 / 108 ( 2013 .01 ) ; AOIK 2217 /07 (21 ) Appl. No. : 15 /923 , 951 ( 2013 .01 ) ; A01K 2227/ 101 (2013 .01 ) ; AOIK ( 22 ) Filed : Mar. 16 , 2018 2217 /075 ( 2013 .01 ) (57 ) ABSTRACT Related U . S . Application Data Materials and methods for making multiplex gene edits in (62 ) Division of application No . 14 /698 ,561 , filed on Apr. cells and are presented . Further methods include animals 28 , 2015, now abandoned . and methods of making the same . Methods of making ( 60 ) Provisional application No . 61/ 985, 327, filed on Apr. chimeric animals are presented , as well as chimeric animals . 28 , 2014 . Specification includes a Sequence Listing . Patent Application Publication Aug . 23 , 2018 Sheet 1 of 13 US 2018 / 0235194 A1 GENERATION OF HOMOZYGOUS CATTLE EDITED AT ONE ALLELE USING SINGLE EDITS Edit allele , Raise FO to Mate FO enough times Raise F1s to Mate F1 siblings Clone cell , sexual maturity , to produce enough F1 sexualmaturity , to make Implant, Gestate 2 years generation carrying 2 years homozygous KO , 9 months , birth edited allele to mate 9 months of FO with each other Generation Primary Fibroblasts ? ? ? Time, years FIG . -

Beyond Type 1 and Type 2 Diabetes; Why Correct Diabetes Classification Is Important Gabriel I. Uwaifo, MD Dept of Endocrinology
Beyond Type 1 and Type 2 Diabetes; Why Correct Diabetes Classification is Important Gabriel I. Uwaifo, MD, FACP, FTOS, FACE. Dept of Endocrinology, Diabetes, Metabolism and Weight Management, Ochsner Medical Center Objectives To highlight various classification methods of diabetes To highlight the importance and consequences of appropriate diabetes classification To provide suggested processes for diabetes classification in primary care settings and indices for specialty referral Presentation outline 1. Case presentations 2. Diabetes classification; past present and future 3. Diabetes classification; why is it important? 4. Suggested schemas for diabetes classification 5. Case presentation conclusions 6. Summary points and conclusions 3 Demonstrative cases Patient DL is a 56 yr old AA gentleman with a BMI of 24 referred for management of his “type 2 diabetes”. He is on basal bolus insulin with current HBA1c of 8.3. His greatest concern is on account of recent onset progressive neurologic symptoms and gaite unsteadiness Patient CY is a 21 yr old Caucasian lady with BMI of 28 and strong family history of diabetes referred for management of her “type 2 diabetes”. She is unsure if she even has diabetes as she indicates most of the SMBGs are under 160 and her current HBA1 is 6.4 on low dose metformin. Patient DR is a 54 yr old Asian lady with BMI of 36 and long standing “type 2 diabetes”. She has been referred because of poor diabetes control on multiple oral antidiabetics and persistent severe hypertriglyceridemia. Questions; Do all -
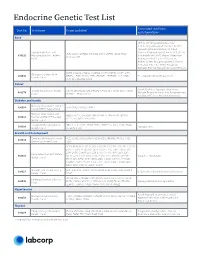
Endocrine Genetic Test List
Endocrine Genetic Test List Associated conditions Test No. Test Name Genes Included and phenotypes Bone HPP, X-Linked Hypophosphatemia, X-Linked Hypophosphatemic Rickets, XLH, Hypophosphatemic Rickets, X-Linked Hypophosphatasia and Dominant Hypophosphatemic Rickets (XLHR), ALPL, CLCN5, CYP2R1, CYP27B1, DMP1, ENPP1, FGF23, PHEX, 630292 Hypophosphatemic Rickets X-Linked Rickets (XLR), Vitamin D-Resistant SLC34A3, VDR Panel Rickets, X-Linked Vitamin D-Resistant Rickets (VDRR), Hypophosphatemic Vitamin D-Resistant Rickets (HPDR), Phosphate Diabetes, Familial Hypophosphatemic Rickets BMP1, COL1A1, COL1A2, CREB3L1, CRTAP, FKBP10, IFITM5, LRP5, Osteogenesis Imperfecta 630543 MBTPS2, P3H1, PLOD2, PPIB, SERPINF1, SERPINH1, SP7, SPARC, OI, Juvenile Primary Osteoporosis Genetic Panel TENT5A, TMEM38B, WNT1 Cancer Familial Isolated Hyperparathyroidism, VistaSeq® Endocrine Cancer CDC73, MAX, MEN1, NF1, PRKAR1A, PTEN, RET, SDHB, SDHC, SDHD, 481374 Multiple Endocrine Neoplasia, Paraganglioma, Panel* TMEM127, TP53 and VHL Parathyroid Cancer, Pheochromocytoma Diabetes and Insulin Maturity-Onset Diabetes of the 630568 GCK, HNF1A, HNF1B, HNF4A Young (MODY) 4-gene Panel Maturity-Onset Diabetes of ABCC8, APPL1, BLK, GCK, HNF1A, HNF1B, HNF4A, INS, KCNJ11, 630513 the Young (MODY) Expanded KLF11, NEUROD1, PAX4, PDX1 Genetic Panel Congenital Hyperinsulinism ABCC8, GCK, GLUD1, HADH, HNF1A, HNF4A, KCNJ11, PGM1, PMM2, 630500 Hypoglycemia Genetic Panel SLC16A1, UCP2 Growth and Development Combined Pituitary Hormone GLI1, HESX1, LHX3, LHX4, OTX2, POU1F1, PROKR2, PROP1, -

An Interesting Case of Young Onset Diabetes Mellitus
International Journal of Research in Medical Sciences Gulati S et al. Int J Res Med Sci. 2017 Sep;5(9):4178-4180 www.msjonline.org pISSN 2320-6071 | eISSN 2320-6012 DOI: http://dx.doi.org/10.18203/2320-6012.ijrms20174007 Case Report An interesting case of young onset diabetes mellitus Shipra Gulati*, Tanvi Batra, Akshay A. Dhamne, Vijayashree S. Gokhale Department of Medicine, Dr. DY Patil Medical College and Hospital, Pimpri, Pune, Maharashtra, India Received: 23 June 2017 Accepted: 22 July 2017 *Correspondence: Dr. Shipra Gulati, E-mail: [email protected] Copyright: © the author(s), publisher and licensee Medip Academy. This is an open-access article distributed under the terms of the Creative Commons Attribution Non-Commercial License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited. ABSTRACT A 24 years old female, was admitted with symptoms of urinary tract infection. She was married and had bad obstetric history. She was known diabetic for 16 years of age and was on regular treatment with injection human insulin mixtard since the time of diagnosis, but had no episode of diabetic ketosis/ ketoacidosis. She had a positive family history of diabetes. She was further evaluated and was found to have normal C peptide levels and islet cell antibodies were found to be negative. Hence, the possibility of MODY (monogenic diabetes) was considered. Her genetic testing could not be done due to financial constraints. But a trial of sulfonylureas was given along with reduction in the dose of insulin to which she responded well and is presently well controlled. -

Guidelines and Recommendations for Laboratory Analysis in the Diagnosis and Management of Diabetes Mellitus
Reviews/Commentaries/ADA Statements POSITION STATEMENT Guidelines and Recommendations for Laboratory Analysis in the Diagnosis and Management of Diabetes Mellitus 1 6 DAVID B. SACKS M. SUE KIRKMAN mellitus, formerly known as insulin- 2 7 MARK ARNOLD AKE LERNMARK 3 8 dependent diabetes mellitus (IDDM) or GEORGE L. BAKRIS BOYD E. METZGER 4 9 juvenile-onset diabetes mellitus, is usu- DAVID E. BRUNS DAVID M. NATHAN 5 ally caused by autoimmune destruction ANDREA RITA HORVATH of the pancreatic islet b-cells, rendering the pancreas unable to synthesize and se- crete insulin (2). Type 2 diabetes mellitus, BACKGROUND—Multiple laboratory tests are used to diagnose and manage patients with diabetes mellitus. The quality of the scientific evidence supporting the use of these tests varies formerly known as non-IDDM or adult- substantially. onset diabetes, is caused by a combina- tion of insulin resistance and inadequate APPROACH—An expert committee compiled evidence-based recommendations for the use of insulin secretion (3,4). Gestational diabe- laboratory testing for patients with diabetes. A new system was developed to grade the overall quality tes mellitus (GDM), which resembles type of the evidence and the strength of the recommendations. Draft guidelines were posted on the fi 2diabetesmorethantype1,develops Internet and presented at the 2007 Arnold O. Beckman Conference. The document was modi ed in during approximately 7% (range, 5%– response to oral and written comments, and a revised draft was posted in 2010 and again modified in response to written comments. The National Academy of Clinical Biochemistry and the Evidence- 15%) of pregnancies, usually remits after Based Laboratory Medicine Committee of the American Association for Clinical Chemistry jointly delivery, and constitutes a major risk fac- reviewed the guidelines, which were accepted after revisions by the Professional Practice Committee tor for the development of type 2 diabetes and subsequently approved by the Executive Committee of the American Diabetes Association. -

Abstracts of Lectures Gfh ÖGH SGMG Tagungsband Abstracts
patients were significantly different across pop mosomal level, but not generally at both levels. Abstracts of Lectures ulations with frequency maximum of the com In this view, the aneuploid karyotype is the read mon mutations in EastEurope (W151X, V326L), out of an underlying chromosomal instability NorthWestEurope (IVS81G>C), and SouthEu (CIN). In a small proportion of cancers display 1. Symposia rope (T93M). ing CIN the loss of this checkpoint is associated Carrier frequency analysis of the IVS81G>C, with the mutational inactivation of a human ho W151X, T93M, and V326L mutations in 2250 mologue of the yeast BUB1 gene. BUB1 controls S1 healthy individuals from different European pop mitotic checkpoints and chromosome segrega ulations revealed much higher frequencies for tion in yeast. The Human SHOX Mutation Database these common mutations (e.g. 1:50 for the IVS8 Because the MIN and CIN forms of instability are Beate Niesler, Christine Fischer and Gudrun A. 1G>C in Austria, and 1:84 for the W151X in rarely found to coexist in tumours, it would seem Rappold Poland) than expected from the reported preva that one form of instability is sufficient to drive Institute of Human Genetics, University of lence of the SLOS. Based on these frequencies tumorigenesis. Heidelberg, Im Neuenheimer Feld 328, the expected incidence of SLOS patients with Genetic instability appears early in tumorigene 69120 Heidelberg, Germany null mutations ranges from 1:2000 to 1:16.000. sis and is believed to play a critical role in the The human SHOX gene (Short Stature Home This discrepancy might be due to underdiagno malignant process. -

Of Analyses January 2019 Imprint
Index of Analyses January 2019 Imprint We reserve the right for errors and alterations. Our laboratory is part of the SYNLAB group. © 3rd edition By SYNLAB Medizinisches Versorgungszentrum Humane Genetik Medical Practice and Laboratory for Human genetics 2 Medical Director Dr. med. Dr. rer. nat. Claudia Nevinny-Stickel-Hinzpeter Consultant Human geneticist Postal Address Lindwurmstrasse 23 D-80337 Munich / Germany Telephone and Fax Reception +49 (0)89. 54 86 29 -0 Invoicing -0 Fax -243 Molecular Genetics -554 Cytogenetics -559 Office hours Monday – Friday 8.30 – 18.00 [email protected] www.humane-genetik.de 3 Table of contents Preanalytics 5 Molecular genetic analyses 8 Cardiac diseases 8 Complex syndromes 13 Connective Tissue Disorders 23 Endocrinology 26 Eye diseases 29 Fertility disorders 30 Gastrointestinal diseases 32 Hematology 33 Hemophilia 35 Hereditary cancer syndromes 36 Immune disorders 44 Intersexuality 45 Kidney diseases 46 Liver diseases 48 Lung diseases 50 Metabolic diseases 51 Mitochondrial diseases 61 Neurodegenerative diseases 64 Neuromuscular diseases /Neuropathies 69 Nutrigenetics 74 Pancreatic diseases 75 Periodic fever 76 Pharmacogenetics 80 Short stature 82 Thrombophilia/Atherosclerosis 84 Uniparental disomies 90 Kinship Analyses 91 Cytogenetics and molecular cytogenetics 92 Prenatal chromosome diagnostics 92 Postnatal chromosome diagnostics 94 Molecular cytogenetics 96 Chromosomal microarray diagnostics 99 Quality assurance 100 Index 104 Index gene names 116 4 Preanalytics Testing Material For genetic testing nuclei-containing cells of the patient are required, which are either cultivated or subjected to DNA extraction. Cells can be harvested from peripheral blood, buccal swabs, amniotic fluid, chorionic villi (CVS), or tissue samples. In case of blood collection, no special preparation of the patient, e.g.