Identification of Dioryctria
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Southern Pine Coneworm, Dioryctria Amatella (Hulst) (Insecta: Lepidoptera: Pyralidae)1 James R
EENY325 Southern Pine Coneworm, Dioryctria amatella (Hulst) (Insecta: Lepidoptera: Pyralidae)1 James R. Meeker2 The Featured Creatures collection provides in-depth profiles pitch masses are larger, appear irregularly-shaped, and flow of insects, nematodes, arachnids and other organisms for months. BTB pitch masses are typically less than 25 relevant to Florida. These profiles are intended for the use of mm in diameter, have an obvious entrance hole, solidify in interested laypersons with some knowledge of biology as well weeks, and are concentrated on the lower bole of large trees as academic audiences. (Barnard and Dixon 1983; Goolsby et al. 1972). Introduction The southern pine coneworm, Dioryctria amatella (Hulst), also commonly referred to as a pitch moth, is consistently one of the most damaging insect pests of pine seed orchard crops throughout the southeastern United States (Ebel et al. 1980). Less well-recognized is that this widespread insect also attacks other parts of pines (Pinus spp.) besides cones. Caterpillars can be found feeding on and in buds, male and female flowers, shoots, branches and stems of all ages and sizes, as well as in conelets (first-year cones) and second- year cones (Ebel 1965; Ebel et al. 1980; Goolsby et al. 1972). The prevalence of D. amatella infestations on forest and Figure 1. Adult and larva of the southern pine coneworm, Dioryctria amatella (Hulst), on a loblolly pinecone. shade trees pines periodically generates concern over the Credits: R. Scott Cameron, International Paper, www.forestryimages. damage it causes. The most noticeable symptom of an org infestation is large masses of pitch exuding from the feeding In addition to cones, susceptible host material includes: sites of caterpillars, hence the name “pitch moth.” Reddish- trees under stress, mechanically injured stems or branches, brown frass may also be evident at feeding sites and is elongating shoots of long leaf (P. -

Lepidoptera of North America 5
Lepidoptera of North America 5. Contributions to the Knowledge of Southern West Virginia Lepidoptera Contributions of the C.P. Gillette Museum of Arthropod Diversity Colorado State University Lepidoptera of North America 5. Contributions to the Knowledge of Southern West Virginia Lepidoptera by Valerio Albu, 1411 E. Sweetbriar Drive Fresno, CA 93720 and Eric Metzler, 1241 Kildale Square North Columbus, OH 43229 April 30, 2004 Contributions of the C.P. Gillette Museum of Arthropod Diversity Colorado State University Cover illustration: Blueberry Sphinx (Paonias astylus (Drury)], an eastern endemic. Photo by Valeriu Albu. ISBN 1084-8819 This publication and others in the series may be ordered from the C.P. Gillette Museum of Arthropod Diversity, Department of Bioagricultural Sciences and Pest Management Colorado State University, Fort Collins, CO 80523 Abstract A list of 1531 species ofLepidoptera is presented, collected over 15 years (1988 to 2002), in eleven southern West Virginia counties. A variety of collecting methods was used, including netting, light attracting, light trapping and pheromone trapping. The specimens were identified by the currently available pictorial sources and determination keys. Many were also sent to specialists for confirmation or identification. The majority of the data was from Kanawha County, reflecting the area of more intensive sampling effort by the senior author. This imbalance of data between Kanawha County and other counties should even out with further sampling of the area. Key Words: Appalachian Mountains, -

GIS Handbook Appendices
Aerial Survey GIS Handbook Appendix D Revised 11/19/2007 Appendix D Cooperating Agency Codes The following table lists the aerial survey cooperating agencies and codes to be used in the agency1, agency2, agency3 fields of the flown/not flown coverages. The contents of this list is available in digital form (.dbf) at the following website: http://www.fs.fed.us/foresthealth/publications/id/id_guidelines.html 28 Aerial Survey GIS Handbook Appendix D Revised 11/19/2007 Code Agency Name AFC Alabama Forestry Commission ADNR Alaska Department of Natural Resources AZFH Arizona Forest Health Program, University of Arizona AZS Arizona State Land Department ARFC Arkansas Forestry Commission CDF California Department of Forestry CSFS Colorado State Forest Service CTAES Connecticut Agricultural Experiment Station DEDA Delaware Department of Agriculture FDOF Florida Division of Forestry FTA Fort Apache Indian Reservation GFC Georgia Forestry Commission HOA Hopi Indian Reservation IDL Idaho Department of Lands INDNR Indiana Department of Natural Resources IADNR Iowa Department of Natural Resources KDF Kentucky Division of Forestry LDAF Louisiana Department of Agriculture and Forestry MEFS Maine Forest Service MDDA Maryland Department of Agriculture MADCR Massachusetts Department of Conservation and Recreation MIDNR Michigan Department of Natural Resources MNDNR Minnesota Department of Natural Resources MFC Mississippi Forestry Commission MODC Missouri Department of Conservation NAO Navajo Area Indian Reservation NDCNR Nevada Department of Conservation -

Lepidoptera, Pyralidae) New to Korea
Anim. Syst. Evol. Divers. Vol. 31, No. 1: 46-50, January 2015 http://dx.doi.org/10.5635/ASED.2015.31.1.046 Short communication Two Species of Phycitinae (Lepidoptera, Pyralidae) New to Korea Mujie Qi, Yang-Seop Bae* Bio-Resource and Environmental Center, College of Life Sciences and Bioengineering, Incheon National University, Incheon 406-772, Korea ABSTRACT Two species of Phycitinae, Rabiria rufimaculella (Yamanaka, 1993) and Copamyntis martimella Kirpichnikova & Yamanaka, 2002, are reported for the first time from Korea. Rabiria rufimaculella can be recognized by having two reddish-yellow and short bands near the postmedial and antemedial line, and by the bifurcate gnathos and the cornutus which is formed by numerous thorn-shaped sclerites in male genitalia. Copamyntis martimella can be distinguished with the congeners by the uniformly distributed setae on the sacculus and the curved aedeagus in male genitalia and the peanut-shaped signum near the middle of the corpus bursae in female genitalia. The adults and genitalia of the species are redescribed and illustrated. Keywords: Pyralidae, Phycitinae, Rabiria, Copamyntis, new records, Korea INTRODUCTION SYSTEMATIC ACCOUNTS The Phycitinae are one of the largest subfamilies of the family Order Lepidoptera Linnaeus, 1758 Pyralidae in Lepidoptera, comprising approximately 5,000 Family Pyralidae Latreille, 1809 species in the world (Li and Ren, 2009). Leech and South Subfamily Phycitinae Ragonot, 1885 (1901) first reported 3 species of Phycitini from the Korean Genus Rabiria Heinrich, 1956 Peninsula; Okamoto (1924), Shibuya (1927), Park and Lee Rabiria Heinrich, 1956: 311. TS: Microphycita conops (1958), Park (1976, 1983, 1993), Byun et al. (1997), Choi et Dyar, 1914. -

Pine Shoot Insects Common in British Columbia
Environment Environnement 1+ Canada Canada Pine Shoot Insects Canadian Service Forestry canadien des Common in Serv~e forits British Columbia David Evans Pacific Forest Research Centre Victoria, British Columbia BC-X-233 PACIFIC FOREST RESEARCH CENTRE The Pacific Forest Research Centre (PFRC) is one of six regional research estab lishments of the Canadian Forestry Service of Environment Canada. The centre conducts a program of work directed toward the solution of major forestry problems and the development of more effective forest management techniques for use in British Columbia and the Yukon. The 30 research projects and over 150 studies which make up the research pro gram of PFRC are divided into three areas known as Forest Protection, Forest Environment and Forest Resources. These are supported by an Economics, Information and Administrative section and a number of specialized research support services such as analytical chemistry, computing microtechnique and remote sensing. Current research projecu, which focus on the establishment, growth and protection of the foresu, include: forest pathology problems, researd1 0f1 seed and cone insecU and disease, biological control of forest pesU, pest damage monitoring and assessment, seed and tree improvement, regenera tion and stand management. ISSN.()705·3274 Pine Shoot Insects Common in British Columbia David Evans Pacific Forest Research Centre 506 Wen Burnside Road. Victoria, British Columbia vaz 1M5 BC·X·233 1982 2 ABSTRACT RESUME This publication is an aid to the identification Ce document aidera a identifier les insectes of pine shoot insects on pines native to British Co des pousses du Pin sur les pins indigenes du Colombie lumbia. -

Reçine Kelebeği Dioryctria Sylvestrella
DOI:http://dx.doi.org/10.16969/teb.75163 Türk. entomol. bült., 2016, 6 (2): 131-141 ISSN 2146-975X Orijinal ara ştırma (Original article) Reçine kelebe ği Dioryctria sylvestrella (Ratzeburg) (Lepidoptera: Pyralidae)’nın Göller Bölgesi ormanlarında zararı, biyolojisi ve do ğal dü şmanları 1 Damage, biology and natural enemies of pine stem borer Dioryctria sylvestrella (Ratzeburg) (Lepidoptera: Pyralidae) in Lake’s District forests Melike B İLENER 2 Mustafa AVCI 2* Summary Dioryctria sylvestrella is a pest that causes significant damage on the brutian pine forests of the Lake’s District, plantation sites. The morphology of this insect, the damage it caused under the field conditions in the forests of the Lake’s District, and its biology were explored while its natural enemies that were effective on the population were determined. They were found mainly on the lower parts of the tree trunks and led to the intensive release of resin. It was observed to have one generation in a year. The larval development of the insects took about eleven months and thus they overwintered as larvae. Starting from the mid-May, the pupae develop inside the resin released and the pupal development took three weeks, the adults flew throughout June depending on the elevation and the climatic conditions. The young larvae emerged from the eggs in late June. The larvae were observed to start feeding first on the barks of the trunk. Throughout this study. Forficula auricularia (Dermaptera: Forficulidae) and Raphidia ophiopsis (Raphidioptera: Raphidiidae) fed with the larvae of D. sylvestrella were observed as the predatory species. As larval and pupal parasitoid, Brachymeria tibialis (Hymenoptera: Chalcididae) and Venturia robusta (Hymenoptera: Ichneumonidae) were determined. -

Shade Tree Borers No
I N S E C T S E R I E S TREES & SHRUBS Shade Tree Borers no. 5.530 by W.S. Cranshaw and D.A. Leatherman 1 Shade tree borers are insects that develop underneath the bark of woody plants. Most of these insects can attack only dying trees, felled logs or trees under stress. Stress to woody plants may be the result of mechanical injury, recent transplanting, overwatering or drought. These borers often are incorrectly blamed Quick Facts... for damage caused by a pre-existing condition or injury. Certain borers, in particular the “clear-wing borers,” are capable of damaging apparently healthy trees. Shade tree borers are insects that develop underneath the Life History and Habits bark of trees and shrubs. Certain A large number of beetles and moths develop as wood borers in their beetles and moths are the most immature (larval) stage. When full-grown, typically in one to two years, the adult common borers. stages cut a hole through the bark and emerge. Many of the adult borers, particularly the longhorned beetles and Most shade tree borers can metallic wood borers, feed on pollen, tender bark or leaves but do not cause any successfully attack only trees significant injury. The adult stage is mostly spent flying to new host trees, mating that are injured or stressed. and laying eggs. Eggs of most shade tree borers are laid on the bark, usually within small Shade tree borer development cracks. Longhorned beetles and horntails deposit their eggs underneath bark. takes from one to three years to Eggs typically hatch within one to two weeks, and the newly emerged complete. -

52 Southern Forest Insect Work Conference
Proceedings 52nd Southern Forest Insect Work Conference Gulfport July 28 – 31, 2009 Courtyard by Marriott Gulfport Beachfront Gulfport, Mississippi PROCEEDINGS 52nd Annual SOUTHERN FOREST INSECT WORK CONFERENCE Courtyard by Marriott Gulfport Beachfront Gulfport, Mississippi 28–31 July 2009 John Nowak, Program Chairman Andy Londo and John Riggins, Local Arrangements Officers: 2008–2009 Chairman ...................................................................................... Scott Salom (2007–2009) Secretary-Treasurer ........................................................................................ Will Shepherd Counselors..................................................................................... Laurie Reid (2005–2009) ....................................................................................... John Nowak (2007–2010) ....................................................................................... Andy Londo (2008–2012) ii TABLE OF CONTENTS Registration List ..................................................................................................................1 Group Pictures .....................................................................................................................2 Program ................................................................................................................................6 Minutes 2009 .....................................................................................................................26 Treasurer's Report .............................................................................................................31 -

Yponomeuta Malinellus
Yponomeuta malinellus Scientific Name Yponomeuta malinellus (Zeller) Synonyms: Hyponomeuta malinella Zeller Hyponomeuta malinellus Zeller Yponomeuta malinella Yponomeuta padella (L.) Yponomeuta padellus malinellus Common Names Apple ermine moth, small ermine moth Figure 1. Y. malinellus adult (Image courtesy of Eric LaGasa, Washington State Department of Agriculture, Bugwood.org). Type of Pest Caterpillar Taxonomic Position Class: Insecta, Order: Lepidoptera, Family: Yponomeutidae Reason for Inclusion 2012 CAPS Additional Pests of Concern Pest Description Eggs: “The individual egg has the appearance of a flattened, yellow, soft disc with the centre area slightly raised, and marked with longitudinal ribbings. Ten to eighty eggs are deposited in overlapping rows to form a flattened, slightly convex, oval egg mass. At the time of deposition, the egg mass is covered with a glutinous substance, which on exposure to air forms a resistant, protective coating. This coating not only acts as an egg-shield but provides an ideal overwintering site for the diapausing first-instar larvae. The egg mass is yellow at first but then darkens until eventually it is grey-brown and resembles the bark of apple twigs. Egg masses average 3-10 mm [0.12-0.39 in] in length and 4 mm [0.16 in] in width but vary considerably in size and shape” (CFIA, 2006). Larvae: “Grey, yellowish-grey, greenish-brown, and greyish-green larvae have been reported. The mature larva is approximately 15-20 mm [0.59-0.79 in] in length; the anterior and posterior extremities are much narrower than the remainder of the body. There are 2 conspicuous laterodorsal black dots on each segment from the mesothorax to the 8th abdominal segment. -
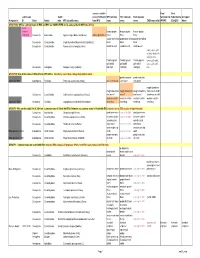
Common Names Working-2008Jan
correct scientific Final Final submission Subfa name (if different WFI common ESC common ESA common Submitted to Submitted to Accepted Assigned to: ID Order Family mily WFI scientific name from WFI) name name name CABI name info? WFIWC: ESA/ESC: Name GROUP 000: Official common name in ESA and ESC are COMPLETED and is same as that in WFI (take off list) need change? bronze poplar bronze poplar bronze poplar 1026-Bup Coleoptera Buprestidae Agrilus liragus Barter and Brown Agrilus granulatus borer borer borer poplar-and-willow poplar-and-willow poplar-and-willow Coleoptera Curculionidae Cryptorhynchus [Sternochetus] lapathi (L.) borer borer borer Coleoptera Curculionidae Nemocestes incomptus (Horn) woods weevil woods weevil wood weevil cooley spruce gall adelgid; douglas fir adelges; sitka Cooley spruce Cooley spruce Cooley spruce spruce gall aphid; gall aphid gall aphid gall aphid spruce gall aphid, Homoptera Adelgidae Adelges cooleyi (Gillette) (adelgid) (adelgid) (adelgid) blue GROUP 00: One of the names in ESA, ESC or WFI differs (Decide by case; likely change highlighted name) ponderosa pine ponderosa pine approved 1989 Lepidoptera Pyralidae Dioryctria auranticella (Grote) pine coneworm coneworm coneworm rough strawberry rough strawberry rough strawberry rough strawberry root weevil; rough Coleoptera Curculionidae Otiorhynchus rugosostriatus (Goeze) root weevil weevil root weevil strawberry weevil western conifer western conifer- western conifer- western conifer- approved 1989 Hemiptera Coreidae Leptoglossus occidentalis Heidemann seed -

Forest Insect Conditions in the United States 1966
FOREST INSECT CONDITIONS IN THE UNITED STATES 1966 FOREST SERVICE ' U.S. DEPARTMENT OF AGRICULTURE Foreword This report is the 18th annual account of the scope, severity, and trend of the more important forest insect infestations in the United States, and of the programs undertaken to check resulting damage and loss. It is compiled primarily for managers of public and private forest lands, but has become useful to students and others interested in outbreak trends and in the location and extent of pest populations. The report also makes possible n greater awareness of the insect prob lem and of losses to the timber resource. The opening section highlights the more important conditions Nationwide, and each section that pertains to a forest region is prefaced by its own brief summary. Under the Federal Forest Pest Control Act, a sharing by Federal and State Governments the costs of surveys and control is resulting in a stronger program of forest insect and disease detection and evaluation surveys on non-Federal lands. As more States avail themselves of this financial assistance from the Federal Government, damage and loss from forest insects will become less. The screening and testing of nonpersistent pesticides for use in suppressing forest defoliators continued in 1966. The carbamate insecticide Zectran in a pilot study of its effectiveness against the spruce budworm in Montana and Idaho appeared both successful and safe. More extensive 'tests are planned for 1967. Since only the smallest of the spray droplets reach the target, plans call for reducing the spray to a fine mist. The course of the fine spray, resulting from diffusion and atmospheric currents, will be tracked by lidar, a radar-laser combination. -

Assessment of Forest Pests and Diseases in Native Boxwood Forests of Georgia Final Report
Assessment of Forest Pests and Diseases in Native Boxwood Forests of Georgia Final report Dr. Iryna Matsiakh Forestry Department, Ukrainian National Forestry University (Lviv) Tbilisi 2016 TABLE OF CONTENT LIST OF TABLES AND FIGURES .................................................................................................................................. 2 ABBREVIATIONS AND ACRONYMS ........................................................................................................................... 5 EXECUTIVE SUMMARY .................................................................................................................................................. 6 INTRODUCTION .............................................................................................................................................................. 10 1. BACKGROUND INFORMATION ............................................................................................................................ 11 1.1. Biodiversity of Georgia ........................................................................................................................................ 11 1.2. Forest Ecosystems .................................................................................................................................................. 12 1.3. Boxwood Forests in Forests Habitat Classification ................................................................................. 14 1.4. Georgian Forests Habitat in the Context of Climate Change