Cloning and Expression of a Cdna Encoding Human Sterol Carrier Protein 2 RITSU YAMAMOTO*, CALEB B
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

SLC44A1 Transport of Choline and Ethanolamine in Disease
SLC44A1 Transport of Choline and Ethanolamine in Disease by Adrian Taylor A Thesis presented to The University of Guelph In partial fulfilment of requirements for the degree of Doctor of Philosophy in Human Health and Nutritional Sciences Guelph, Ontario, Canada © Adrian Taylor, April, 2019 ABSTRACT SLC44A1 TRANSPORT OF CHOLINE AND ETHANOLAMINE IN DISEASE Adrian Taylor Advisor(s): University of Guelph, 2019 Marica Bakovic Choline and ethanolamine are important molecules required for the de novo synthesis of phosphatidylcholine (PC) and phosphatidylethanolamine (PE) via the Kennedy pathway. Additionally, these two molecules are vital for maintaining both muscular and neurological function. The goal of this thesis was to gain insight into PC and PE metabolism with the use of unique metabolic disturbances ranging from obesity and genetic mutations in neurodegenerative disease. Firstly, the protective effects of choline supplementation on muscular function were investigated within the Pcyt2+/- mouse model. In Pcyt2+/- mice, substrate flow through the CDP-ethanolamine branch of the Kennedy pathway was diminished resulting in triacylglycerol (TAG) accumulation and obesity. Supplemental choline improved muscle function by altering the expression of genes devoted to reducing TAG synthesis and restoring energy homeostasis. With this new insight about the role of choline in regulating metabolism, the cellular uptake mechanism of choline was then analyzed. Skin fibroblasts from two patients with homozygous mutations in the SLC44A1 gene suffering from Neurodegeneration with Brain Iron Accumulation (NBIA) were utilized. In these fibroblasts, SLC44A1 expression and choline uptake were drastically diminished. Moreover, PC levels were unaffected while PE levels were diminished relative to control, an indication of perturbed phospholipid homeostasis. -

Upregulation of Peroxisome Proliferator-Activated Receptor-Α And
Upregulation of peroxisome proliferator-activated receptor-α and the lipid metabolism pathway promotes carcinogenesis of ampullary cancer Chih-Yang Wang, Ying-Jui Chao, Yi-Ling Chen, Tzu-Wen Wang, Nam Nhut Phan, Hui-Ping Hsu, Yan-Shen Shan, Ming-Derg Lai 1 Supplementary Table 1. Demographics and clinical outcomes of five patients with ampullary cancer Time of Tumor Time to Age Differentia survival/ Sex Staging size Morphology Recurrence recurrence Condition (years) tion expired (cm) (months) (months) T2N0, 51 F 211 Polypoid Unknown No -- Survived 193 stage Ib T2N0, 2.41.5 58 F Mixed Good Yes 14 Expired 17 stage Ib 0.6 T3N0, 4.53.5 68 M Polypoid Good No -- Survived 162 stage IIA 1.2 T3N0, 66 M 110.8 Ulcerative Good Yes 64 Expired 227 stage IIA T3N0, 60 M 21.81 Mixed Moderate Yes 5.6 Expired 16.7 stage IIA 2 Supplementary Table 2. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of an ampullary cancer microarray using the Database for Annotation, Visualization and Integrated Discovery (DAVID). This table contains only pathways with p values that ranged 0.0001~0.05. KEGG Pathway p value Genes Pentose and 1.50E-04 UGT1A6, CRYL1, UGT1A8, AKR1B1, UGT2B11, UGT2A3, glucuronate UGT2B10, UGT2B7, XYLB interconversions Drug metabolism 1.63E-04 CYP3A4, XDH, UGT1A6, CYP3A5, CES2, CYP3A7, UGT1A8, NAT2, UGT2B11, DPYD, UGT2A3, UGT2B10, UGT2B7 Maturity-onset 2.43E-04 HNF1A, HNF4A, SLC2A2, PKLR, NEUROD1, HNF4G, diabetes of the PDX1, NR5A2, NKX2-2 young Starch and sucrose 6.03E-04 GBA3, UGT1A6, G6PC, UGT1A8, ENPP3, MGAM, SI, metabolism -
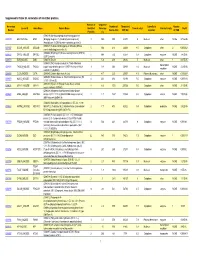
Supporting Table S3 for PDF Maker
Supplemental Table S3. Annotation of identified proteins. Number of Sequence Accession Number of Theoretical Subcellular Number Locus ID Gene Name Protein Name Identified Coverage Theoretical pI Protein Family NSAF Number Amino Acid MW (Da) Location of TMD Peptides (%) (O08539) Myc box-dependent-interacting protein 1 O08539 BIN1_MOUSE BIN1 (Bridging integrator 1) (Amphiphysin-like protein) 3 10.5 588 64470 5 Nucleus other NONE 5.73E-05 (Amphiphysin II) (SH3-domain-containing protein 9) (O08547) Vesicle-trafficking protein SEC22b (SEC22 O08547 SC22B_MOUSE SEC22B 5 30.4 214 24609 8.5 Cytoplasm other 2 0.000262 vesicle-trafficking protein-like 1) (O08553) Dihydropyrimidinase-related protein 2 (DRP-2) O08553 DPYL2_MOUSE DPYSL2 5 19.4 572 62278 6.4 Cytoplasm enzyme NONE 7.85E-05 (ULIP 2 protein) O08579 EMD_MOUSE EMD (O08579) Emerin 1 5.8 259 29436 5 Nucleus other 1 8.67E-05 (O08583) THO complex subunit 4 (Tho4) (RNA and transcription O08583 THOC4_MOUSE THOC4 export factor-binding protein 1) (REF1-I) (Ally of AML-1 1 9.8 254 26809 11.2 Nucleus NONE 2.21E-05 regulator and LEF-1) (Aly/REF) O08585 CLCA_MOUSE CLTA (O08585) Clathrin light chain A (Lca) 2 4.7 235 25557 4.5 Plasma Membrane other NONE 0.000287 (O08600) Endonuclease G, mitochondrial precursor (EC O08600 NUCG_MOUSE ENDOG 4 23.1 294 32191 9.5 Cytoplasm enzyme NONE 0.000134 3.1.30.-) (Endo G) (O08638) Myosin-11 (Myosin heavy chain, smooth O08638 MYH11_MOUSE MYH11 8 6.6 1972 227026 5.5 Cytoplasm other NONE 3.13E-05 muscle isoform) (SMMHC) (O08648) Mitogen-activated protein kinase kinase -

KONSTRUKTION VON ESCHERICHIA COLI PRODUKTIONSSTÄMMEN ZUR FERMENTATIVEN HERSTELLUNG VON SUCCINAT AUS GLYCERIN Stefan Söllner
KONSTRUKTION VON ESCHERICHIA COLI PRODUKTIONSSTÄMMEN ZUR FERMENTATIVEN HERSTELLUNG VON SUCCINAT AUS GLYCERIN Von der Fakultät 4 (Energie-, Verfahrens- und Biotechnik) der Universität Stuttgart zur Erlangung der Würde eines Doktors der Naturwissenschaften (Dr. rer. nat.) genehmigte Abhandlung Vorgelegt von Stefan Söllner aus Schweinfurt Hauptberichter: Prof. Dr. rer. nat. Ralf Mattes Mitberichter: Prof. Dr.-Ing. Ralf Takors Tag der mündlichen Prüfung: 29.02.2012 Institut für Industrielle Genetik Universität Stuttgart 2012 Vielen Dank! … Herrn Prof. Dr. R. Mattes danke ich für die Überlassung des interessanten Themas, des Arbeitsplatzes, für gelegentliches Aufmuntern und für die Erstellung des Erstgutachtens dieser Arbeit. … Herrn Prof. Dr. R. Takors danke ich für die freundliche Übernahme des Zweitgutachtens und für spannende Diskussionen. … Herrn Dr. Josef Altenbuchner danke ich für die praktische Betreuung dieser Arbeit, für diverse Einladungen zu Grillfesten und ganz besonders für die intensive Durchsicht des Manuskriptes!!! … Herrn Dr. Martin Siemann-Herzberg danke ich für die motivierende, überschwängliche Begeisterung, die meine Ideen und Ergebnisse bei deren Besprechung jedesmal auslösten. … Herrn Prof. Dr. Reuss danke ich für die Initiierung des Projektes lange vor meiner Zeit. … Meinen Kollegen und Exkollegen danke ich für die gute Zusammenarbeit, das abwechslungsreiche Arbeitsklima sowie die vielen fachlichen und nichtfachlichen Gespräche, welche die Arbeit immer spannend gestalteten. Vor allem danke ich für das Verständnis für die von mir durchgeführten, absolut notwendigen, regelmäßigen Arbeitskontrollen. … Frau Dr. Anne Völker hat mir die Integration zu Beginn meines Aufenthaltes am IIG sehr erleichtert. Herzlichen Dank dafür! … Herrn Kambiz Morabbi Heravi danke ich recht herzlich für die Einladung ans National Institute of Genetic Engineering and Biotechnology in Teheran, Iran und für die internationale Freundschaft. -

Discovery of Novel Biomarkers for Atherosclerotic Aortic Aneurysm
www.nature.com/scientificreports There are amendments to this paper OPEN Discovery of novel biomarkers for atherosclerotic aortic aneurysm through proteomics-based assessment of disease progression Hiroaki Yagi1,6, Mitsuhiro Nishigori1,2,6, Yusuke Murakami2,6, Tsukasa Osaki1, Sayaka Muto2,4, Yutaka Iba3, Kenji Minatoya3, Yoshihiko Ikeda4, Hatsue Ishibashi-Ueda4, Takayuki Morisaki5, Hitoshi Ogino3, Hiroshi Tanaka3, Hiroaki Sasaki3, Hitoshi Matsuda3 & Naoto Minamino1,2* Since aortic aneurysms (AAs) are mostly asymptomatic, but they have a high mortality rate upon rupture, their detection and progression evaluation are clinically important issues. To discover diagnostic biomarkers for AA, we performed proteome analysis of aortic media from patients with thoracic atherosclerotic AA (TAAA), comparing protein levels between the aneurysm and normal tissue areas. After hierarchical clustering analysis of the proteome analysis data, tissue samples were classifed into three groups, regardless of morphological features. This classifcation was shown to refect disease progression stage identifed by pathological examination. This proteomics-based staging system enabled us to identify more signifcantly altered proteins than the morphological classifcation system. In subsequent data analysis, Niemann-Pick disease type C2 protein (NPC2) and insulin-like growth factor-binding protein 7 (IGFBP7) were selected as novel biomarker candidates for AA and were compared with the previously reported biomarker, thrombospondin 1 (THBS1). Blood concentrations of NPC2 and IGFBP7 were signifcantly increased, while THBS1 levels were decreased in TAAA and abdominal atherosclerotic AA patients. Receiver operating characteristic analysis of AA patients and healthy controls showed that NPC2 and IGFBP7 have higher specifcity and sensitivity than THBS1. Thus, NPC2 and IGFBP7 are promising biomarkers for the detection and progression evaluation of AA. -

Proteomics of Lipid Accumulation and DGAT Inhibition in Hepg2 Liver Carcinoma Cells
Proteomics of lipid accumulation and DGAT inhibition in HepG2 liver carcinoma cells. By Bhumika Bhatt-Wessel A thesis submitted to Victoria University of Wellington in fulfilment of the requirement for the degree of Doctor of Philosophy In Cell and Molecular Biology. Victoria University of Wellington 2017. i ABSTRACT Non-alcoholic fatty liver disease (NAFLD) is a manifestation of the metabolic syndrome in the liver. It is marked by hepatocyte accumulation of triacylglycerol (TAG) rich lipid droplets. In some patients, the disease progresses to non-alcoholic steatohepatitis (NASH), characterized by cellular damage, inflammation and fibrosis. In some cases, cirrhosis and liver failure may occur. However, the pathogenesis of NAFLD is still unclear. The present project is based on the hypothesis that hepatocytes are equipped with mechanisms that allow them to manage lipid accumulation to a certain extent. Continued or increased lipid accumulation beyond this triggers molecular mechanisms such as oxidative stress, lipid peroxidation and cell death that aggravate the condition and cause disease progression. The aim of this project is to study the effects of lipid accumulation on the cells using proteomics approach to identify proteins involved in the disease progression. A cell culture model was used in the study. HepG2 cells, a human liver carcinoma cell line, were treated with a mixture of fatty acids (FA) to induce lipid accumulation. The lipid accumulation in HepG2 cells was measured with Oil red O assay and the effect of lipid accumulation on the proliferation of the cells was measured using an MTT cell proliferation assay. HepG2 cells treated with 1 mM FA mixture for 6 hours induced lipid accumulation 1.4 times of control with 90% of cell proliferation capacity of the control cells. -

Fundamental Differences in Cell Cycle Deregulation in Human Papillomavirus–Positive and Human Papillomavirus–Negative Head/Neck and Cervical Cancers
Research Article Fundamental Differences in Cell Cycle Deregulation in Human Papillomavirus–Positive and Human Papillomavirus–Negative Head/Neck and Cervical Cancers Dohun Pyeon,1,2 Michael A. Newton,3 Paul F. Lambert,1 Johan A. den Boon,1,2 Srikumar Sengupta,1,2 Carmen J. Marsit,6 Craig D. Woodworth,7 Joseph P. Connor,4 Thomas H. Haugen,8 Elaine M. Smith,9 Karl T. Kelsey,6 Lubomir P. Turek,8 and Paul Ahlquist1,2,5 1McArdle Laboratory for Cancer Research; 2Institute for Molecular Virology; Departments of 3Statistics and of Biostatistics and Medical Informatics and 4Obstetrics and Gynecology; 5Howard Hughes Medical Institute, University of Wisconsin-Madison, Madison, Wisconsin; 6Departments of Genetics and Complex Diseases, Harvard School of Public Health, Boston, Massachusetts; 7Department of Biology, Clarkson University, Potsdam, New York; 8Department of Pathology, Veterans Affairs Medical Center; and 9Department of Epidemiology, University of Iowa, Iowa City, Iowa Abstract lower female reproductive tract, penis, and anus (1). In particular, Human papillomaviruses (HPV) are associated with nearly all high-risk HPVs are associated with nearly all cervical cancers, a cervical cancers, 20% to 30% of head and neck cancers (HNC), leading cause of cancer death in women worldwide despite the and other cancers. Because HNCs also arise in HPV-negative effectiveness in developed countries of screening for early patients, this type of cancer provides unique opportunities to detection of precancerous lesions (1). Prophylactic HPV vaccines define similarities and differences of HPV-positive versus HPV- should eventually reduce infections by the most prevalent high-risk HPVs, but do not cover all high-risk HPVs. -

Proteomic Survey Towards the Tissue-Specific Proteins of Mouse Mitochondria
SCIENCE CHINA Life Sciences SPECIAL TOPIC January 2011 Vol.54 No.1: 3–15 • RESEARCH PAPERS • doi: 10.1007/s11427-010-4107-0 Proteomic survey towards the tissue-specific proteins of mouse mitochondria WANG Yuan, SUN HaiDan, RU YaWei, YIN SongYue, YIN Liang & LIU SiQi* Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 101300, China Received June 29, 2010; accepted July 30, 2010 Mitochondrion plays the key functions in mammalian cells. It is believed that mitochondrion exerts the common biologic func- tions in many tissues, but also performs some specific functions correspondent with tissues where it is localized. To identify the tissue-specific mitochondrial proteins, we carried out a systematic survey towards mitochondrial proteins in the tissues of C57BL/6J mouse, such as liver, kidney and heart. The mitochondrial proteins were separated by 2DE and identified by MALDI-TOF/TOF MS. Total of 87 unique proteins were identified as the tissue-specific ones, and some representatives were further verified through ICPL quantification and Western blot. Because these issue-specific proteins are coded from nuclear genes, real-time PCR was employed to examine the mRNA status of six typical genes found in the tissues.With combining of the expression data and the co-localization images obtained from confocal microscope, we came to the conclusion that the tis- sue-specifically mitochondrial proteins were widely distributed among the mouse tissues. Our investigation, therefore, indeed provides a solid base to further explore the biological significance of the mitochondrial proteins with tissue-orientation. proteomics, tissue-specific, mitochondria Citation: Wang Y, Sun H D, Ru Y W, et al. -
![Sterol Carrier Protein 2 (SCP2) Mouse Monoclonal Antibody [Clone ID: OTI1E4] Product Data](https://docslib.b-cdn.net/cover/4088/sterol-carrier-protein-2-scp2-mouse-monoclonal-antibody-clone-id-oti1e4-product-data-2034088.webp)
Sterol Carrier Protein 2 (SCP2) Mouse Monoclonal Antibody [Clone ID: OTI1E4] Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for CF507247 Sterol carrier protein 2 (SCP2) Mouse Monoclonal Antibody [Clone ID: OTI1E4] Product data: Product Type: Primary Antibodies Clone Name: OTI1E4 Applications: IHC, WB Recommended Dilution: WB 1:4000, IHC 1:150 Reactivity: Human, Mouse, Rat Host: Mouse Isotype: IgG1 Clonality: Monoclonal Immunogen: Full length human recombinant protein of human SCP2(NP_002970) produced in HEK293T cell. Formulation: Lyophilized powder (original buffer 1X PBS, pH 7.3, 8% trehalose) Reconstitution Method: For reconstitution, we recommend adding 100uL distilled water to a final antibody concentration of about 1 mg/mL. To use this carrier-free antibody for conjugation experiment, we strongly recommend performing another round of desalting process. (OriGene recommends Zeba Spin Desalting Columns, 7KMWCO from Thermo Scientific) Purification: Purified from mouse ascites fluids or tissue culture supernatant by affinity chromatography (protein A/G) Conjugation: Unconjugated Storage: Store at -20°C as received. Stability: Stable for 12 months from date of receipt. Predicted Protein Size: 58.8 kDa Gene Name: Homo sapiens sterol carrier protein 2 (SCP2), transcript variant 1, mRNA. Database Link: NP_002970 Entrez Gene 20280 MouseEntrez Gene 25541 RatEntrez Gene 6342 Human P22307 This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 3 Sterol carrier protein 2 (SCP2) Mouse Monoclonal Antibody [Clone ID: OTI1E4] – CF507247 Background: This gene encodes two proteins: sterol carrier protein X (SCPx) and sterol carrier protein 2 (SCP2), as a result of transcription initiation from 2 independently regulated promoters. -

RNA-Seq Reveals Altered Gene Expression Levels in Proximal
www.nature.com/scientificreports OPEN RNA‑seq reveals altered gene expression levels in proximal tubular cell cultures compared to renal cortex but not during early glucotoxicity Linnéa M. Nilsson1, Miguel Castresana‑Aguirre 2, Lena Scott3 & Hjalmar Brismar 1,3* Cell cultures are often used to study physiological processes in health and disease. It is well‑known that cells change their gene expression in vitro compared to in vivo, but it is rarely experimentally addressed. High glucose is a known trigger of apoptosis in proximal tubular cells (PTC). Here we used RNA-seq to detect diferentially expressed genes in cultures of primary rat PTC, 3 days old, compared to cells retrieved directly from rat outer renal cortex and between PTC exposed to 15 mM glucose and control for 8 h. The expression of 6,174 genes was signifcantly up- or downregulated in the cultures of PTC compared to the cells in the outer renal cortex. Most altered were mitochondrial and metabolism related genes. Gene expression of proapoptotic proteins were upregulated and gene expression of antiapoptotic proteins were downregulated in PTC. Expression of transporter related genes were generally downregulated. After 8 h, high glucose had not altered the gene expression in PTC. The current study provides evidence that cells alter their gene expression in vitro compared to in vivo and suggests that short‑term high glucose exposure can trigger apoptosis in PTC without changing the gene expression levels of apoptotic proteins. Cell cultures, both primary and immortalized, are ofen used as models in biological research to investigate physi- ological processes in health and disease. -

(12) Patent Application Publication (10) Pub. No.: US 2003/0198970 A1 Roberts (43) Pub
US 2003O19897OA1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2003/0198970 A1 Roberts (43) Pub. Date: Oct. 23, 2003 (54) GENOSTICS clinical trials on groups or cohorts of patients. This group data is used to derive a Standardised method of treatment (75) Inventor: Gareth Wyn Roberts, Cambs (GB) which is Subsequently applied on an individual basis. There is considerable evidence that a significant factor underlying Correspondence Address: the individual variability in response to disease, therapy and FINNEGAN, HENDERSON, FARABOW, prognosis lies in a person's genetic make-up. There have GARRETT & DUNNER been numerous examples relating that polymorphisms LLP within a given gene can alter the functionality of the protein 1300 ISTREET, NW encoded by that gene thus leading to a variable physiological WASHINGTON, DC 20005 (US) response. In order to bring about the integration of genomics into medical practice and enable design and building of a (73) Assignee: GENOSTIC PHARMA LIMITED technology platform which will enable the everyday practice (21) Appl. No.: 10/206,568 of molecular medicine a way must be invented for the DNA Sequence data to be aligned with the identification of genes (22) Filed: Jul. 29, 2002 central to the induction, development, progression and out come of disease or physiological States of interest. Accord Related U.S. Application Data ing to the invention, the number of genes and their configu rations (mutations and polymorphisms) needed to be (63) Continuation of application No. 09/325,123, filed on identified in order to provide critical clinical information Jun. 3, 1999, now abandoned. concerning individual prognosis is considerably less than the 100,000 thought to comprise the human genome. -

Metabolic Network-Based Stratification of Hepatocellular Carcinoma Reveals Three Distinct Tumor Subtypes
Metabolic network-based stratification of hepatocellular carcinoma reveals three distinct tumor subtypes Gholamreza Bidkhoria,b,1, Rui Benfeitasa,1, Martina Klevstigc,d, Cheng Zhanga, Jens Nielsene, Mathias Uhlena, Jan Borenc,d, and Adil Mardinoglua,b,e,2 aScience for Life Laboratory, KTH Royal Institute of Technology, SE-17121 Stockholm, Sweden; bCentre for Host-Microbiome Interactions, Dental Institute, King’s College London, SE1 9RT London, United Kingdom; cDepartment of Molecular and Clinical Medicine, University of Gothenburg, SE-41345 Gothenburg, Sweden; dThe Wallenberg Laboratory, Sahlgrenska University Hospital, SE-41345 Gothenburg, Sweden; and eDepartment of Biology and Biological Engineering, Chalmers University of Technology, SE-41296 Gothenburg, Sweden Edited by Sang Yup Lee, Korea Advanced Institute of Science and Technology, Daejeon, Republic of Korea, and approved November 1, 2018 (received for review April 27, 2018) Hepatocellular carcinoma (HCC) is one of the most frequent forms of of markers associated with recurrence and poor prognosis (13–15). liver cancer, and effective treatment methods are limited due to Moreover, genome-scale metabolic models (GEMs), collections tumor heterogeneity. There is a great need for comprehensive of biochemical reactions, and associated enzymes and transporters approaches to stratify HCC patients, gain biological insights into have been successfully used to characterize the metabolism of subtypes, and ultimately identify effective therapeutic targets. We HCC, as well as identify drug targets for HCC patients (11, 16–18). stratified HCC patients and characterized each subtype using tran- For instance, HCC tumors have been stratified based on the uti- scriptomics data, genome-scale metabolic networks and network lization of acetate (11). Analysis of HCC metabolism has also led topology/controllability analysis.