Proteomic Survey Towards the Tissue-Specific Proteins of Mouse Mitochondria
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

SLC44A1 Transport of Choline and Ethanolamine in Disease
SLC44A1 Transport of Choline and Ethanolamine in Disease by Adrian Taylor A Thesis presented to The University of Guelph In partial fulfilment of requirements for the degree of Doctor of Philosophy in Human Health and Nutritional Sciences Guelph, Ontario, Canada © Adrian Taylor, April, 2019 ABSTRACT SLC44A1 TRANSPORT OF CHOLINE AND ETHANOLAMINE IN DISEASE Adrian Taylor Advisor(s): University of Guelph, 2019 Marica Bakovic Choline and ethanolamine are important molecules required for the de novo synthesis of phosphatidylcholine (PC) and phosphatidylethanolamine (PE) via the Kennedy pathway. Additionally, these two molecules are vital for maintaining both muscular and neurological function. The goal of this thesis was to gain insight into PC and PE metabolism with the use of unique metabolic disturbances ranging from obesity and genetic mutations in neurodegenerative disease. Firstly, the protective effects of choline supplementation on muscular function were investigated within the Pcyt2+/- mouse model. In Pcyt2+/- mice, substrate flow through the CDP-ethanolamine branch of the Kennedy pathway was diminished resulting in triacylglycerol (TAG) accumulation and obesity. Supplemental choline improved muscle function by altering the expression of genes devoted to reducing TAG synthesis and restoring energy homeostasis. With this new insight about the role of choline in regulating metabolism, the cellular uptake mechanism of choline was then analyzed. Skin fibroblasts from two patients with homozygous mutations in the SLC44A1 gene suffering from Neurodegeneration with Brain Iron Accumulation (NBIA) were utilized. In these fibroblasts, SLC44A1 expression and choline uptake were drastically diminished. Moreover, PC levels were unaffected while PE levels were diminished relative to control, an indication of perturbed phospholipid homeostasis. -

Upregulation of Peroxisome Proliferator-Activated Receptor-Α And
Upregulation of peroxisome proliferator-activated receptor-α and the lipid metabolism pathway promotes carcinogenesis of ampullary cancer Chih-Yang Wang, Ying-Jui Chao, Yi-Ling Chen, Tzu-Wen Wang, Nam Nhut Phan, Hui-Ping Hsu, Yan-Shen Shan, Ming-Derg Lai 1 Supplementary Table 1. Demographics and clinical outcomes of five patients with ampullary cancer Time of Tumor Time to Age Differentia survival/ Sex Staging size Morphology Recurrence recurrence Condition (years) tion expired (cm) (months) (months) T2N0, 51 F 211 Polypoid Unknown No -- Survived 193 stage Ib T2N0, 2.41.5 58 F Mixed Good Yes 14 Expired 17 stage Ib 0.6 T3N0, 4.53.5 68 M Polypoid Good No -- Survived 162 stage IIA 1.2 T3N0, 66 M 110.8 Ulcerative Good Yes 64 Expired 227 stage IIA T3N0, 60 M 21.81 Mixed Moderate Yes 5.6 Expired 16.7 stage IIA 2 Supplementary Table 2. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of an ampullary cancer microarray using the Database for Annotation, Visualization and Integrated Discovery (DAVID). This table contains only pathways with p values that ranged 0.0001~0.05. KEGG Pathway p value Genes Pentose and 1.50E-04 UGT1A6, CRYL1, UGT1A8, AKR1B1, UGT2B11, UGT2A3, glucuronate UGT2B10, UGT2B7, XYLB interconversions Drug metabolism 1.63E-04 CYP3A4, XDH, UGT1A6, CYP3A5, CES2, CYP3A7, UGT1A8, NAT2, UGT2B11, DPYD, UGT2A3, UGT2B10, UGT2B7 Maturity-onset 2.43E-04 HNF1A, HNF4A, SLC2A2, PKLR, NEUROD1, HNF4G, diabetes of the PDX1, NR5A2, NKX2-2 young Starch and sucrose 6.03E-04 GBA3, UGT1A6, G6PC, UGT1A8, ENPP3, MGAM, SI, metabolism -

Health Effects Support Document for Perfluorooctanoic Acid (PFOA)
United States Office of Water EPA 822-R-16-003 Environmental Protection Mail Code 4304T May 2016 Agency Health Effects Support Document for Perfluorooctanoic Acid (PFOA) Perfluorooctanoic Acid – May 2016 i Health Effects Support Document for Perfluorooctanoic Acid (PFOA) U.S. Environmental Protection Agency Office of Water (4304T) Health and Ecological Criteria Division Washington, DC 20460 EPA Document Number: 822-R-16-003 May 2016 Perfluorooctanoic Acid – May 2016 ii BACKGROUND The Safe Drinking Water Act (SDWA), as amended in 1996, requires the Administrator of the U.S. Environmental Protection Agency (EPA) to periodically publish a list of unregulated chemical contaminants known or anticipated to occur in public water systems and that may require regulation under SDWA. The SDWA also requires the Agency to make regulatory determinations on at least five contaminants on the Contaminant Candidate List (CCL) every 5 years. For each contaminant on the CCL, before EPA makes a regulatory determination, the Agency needs to obtain sufficient data to conduct analyses on the extent to which the contaminant occurs and the risk it poses to populations via drinking water. Ultimately, this information will assist the Agency in determining the most appropriate course of action in relation to the contaminant (e.g., developing a regulation to control it in drinking water, developing guidance, or deciding not to regulate it). The PFOA health assessment was initiated by the Office of Water, Office of Science and Technology in 2009. The draft Health Effects Support Document for Perfluoroctanoic Acid (PFOA) was completed in 2013 and released for public comment in February 2014. -

(LCHAD) Deficiency / Mitochondrial Trifunctional Protein (MTF) Deficiency
Long chain acyl-CoA dehydrogenase (LCHAD) deficiency / Mitochondrial trifunctional protein (MTF) deficiency Contact details Introduction Regional Genetics Service Long chain acyl-CoA dehydrogenase (LCHAD) deficiency / mitochondrial trifunctional Levels 4-6, Barclay House protein (MTF) deficiency is an autosomal recessive disorder of mitochondrial beta- 37 Queen Square oxidation of fatty acids. The mitochondrial trifunctional protein is composed of 4 alpha London, WC1N 3BH and 4 beta subunits, which are encoded by the HADHA and HADHB genes, respectively. It is characterized by early-onset cardiomyopathy, hypoglycemia, T +44 (0) 20 7762 6888 neuropathy, and pigmentary retinopathy, and sudden death. There is also an infantile F +44 (0) 20 7813 8578 onset form with a hepatic Reye-like syndrome, and a late-adolescent onset form with primarily a skeletal myopathy. Tandem mass spectrometry of organic acids in urine, Samples required and carnitines in blood spots, allows the diagnosis to be unequivocally determined. An 5ml venous blood in plastic EDTA additional clinical complication can occur in the pregnant mothers of affected fetuses; bottles (>1ml from neonates) they may experience maternal acute fatty liver of pregnancy (AFLP) syndrome or Prenatal testing must be arranged hypertension/haemolysis, elevated liver enzymes and low platelets (HELLP) in advance, through a Clinical syndrome. Genetics department if possible. The genes encoding the HADHA and HADHB subunits are located on chromosome Amniotic fluid or CV samples 2p23.3. The pathogenic -

Lipid Metabolic Reprogramming: Role in Melanoma Progression and Therapeutic Perspectives
cancers Review Lipid metabolic Reprogramming: Role in Melanoma Progression and Therapeutic Perspectives 1, 1, 1 2 1 Laurence Pellerin y, Lorry Carrié y , Carine Dufau , Laurence Nieto , Bruno Ségui , 1,3 1, , 1, , Thierry Levade , Joëlle Riond * z and Nathalie Andrieu-Abadie * z 1 Centre de Recherches en Cancérologie de Toulouse, Equipe Labellisée Fondation ARC, Université Fédérale de Toulouse Midi-Pyrénées, Université Toulouse III Paul-Sabatier, Inserm 1037, 2 avenue Hubert Curien, tgrCS 53717, 31037 Toulouse CEDEX 1, France; [email protected] (L.P.); [email protected] (L.C.); [email protected] (C.D.); [email protected] (B.S.); [email protected] (T.L.) 2 Institut de Pharmacologie et de Biologie Structurale, CNRS, Université Toulouse III Paul-Sabatier, UMR 5089, 205 Route de Narbonne, 31400 Toulouse, France; [email protected] 3 Laboratoire de Biochimie Métabolique, CHU Toulouse, 31059 Toulouse, France * Correspondence: [email protected] (J.R.); [email protected] (N.A.-A.); Tel.: +33-582-7416-20 (J.R.) These authors contributed equally to this work. y These authors jointly supervised this work. z Received: 15 September 2020; Accepted: 23 October 2020; Published: 27 October 2020 Simple Summary: Melanoma is a devastating skin cancer characterized by an impressive metabolic plasticity. Melanoma cells are able to adapt to the tumor microenvironment by using a variety of fuels that contribute to tumor growth and progression. In this review, the authors summarize the contribution of the lipid metabolic network in melanoma plasticity and aggressiveness, with a particular attention to specific lipid classes such as glycerophospholipids, sphingolipids, sterols and eicosanoids. -
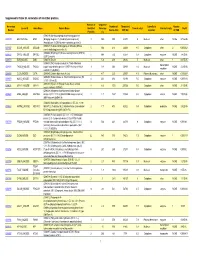
Supporting Table S3 for PDF Maker
Supplemental Table S3. Annotation of identified proteins. Number of Sequence Accession Number of Theoretical Subcellular Number Locus ID Gene Name Protein Name Identified Coverage Theoretical pI Protein Family NSAF Number Amino Acid MW (Da) Location of TMD Peptides (%) (O08539) Myc box-dependent-interacting protein 1 O08539 BIN1_MOUSE BIN1 (Bridging integrator 1) (Amphiphysin-like protein) 3 10.5 588 64470 5 Nucleus other NONE 5.73E-05 (Amphiphysin II) (SH3-domain-containing protein 9) (O08547) Vesicle-trafficking protein SEC22b (SEC22 O08547 SC22B_MOUSE SEC22B 5 30.4 214 24609 8.5 Cytoplasm other 2 0.000262 vesicle-trafficking protein-like 1) (O08553) Dihydropyrimidinase-related protein 2 (DRP-2) O08553 DPYL2_MOUSE DPYSL2 5 19.4 572 62278 6.4 Cytoplasm enzyme NONE 7.85E-05 (ULIP 2 protein) O08579 EMD_MOUSE EMD (O08579) Emerin 1 5.8 259 29436 5 Nucleus other 1 8.67E-05 (O08583) THO complex subunit 4 (Tho4) (RNA and transcription O08583 THOC4_MOUSE THOC4 export factor-binding protein 1) (REF1-I) (Ally of AML-1 1 9.8 254 26809 11.2 Nucleus NONE 2.21E-05 regulator and LEF-1) (Aly/REF) O08585 CLCA_MOUSE CLTA (O08585) Clathrin light chain A (Lca) 2 4.7 235 25557 4.5 Plasma Membrane other NONE 0.000287 (O08600) Endonuclease G, mitochondrial precursor (EC O08600 NUCG_MOUSE ENDOG 4 23.1 294 32191 9.5 Cytoplasm enzyme NONE 0.000134 3.1.30.-) (Endo G) (O08638) Myosin-11 (Myosin heavy chain, smooth O08638 MYH11_MOUSE MYH11 8 6.6 1972 227026 5.5 Cytoplasm other NONE 3.13E-05 muscle isoform) (SMMHC) (O08648) Mitogen-activated protein kinase kinase -

KONSTRUKTION VON ESCHERICHIA COLI PRODUKTIONSSTÄMMEN ZUR FERMENTATIVEN HERSTELLUNG VON SUCCINAT AUS GLYCERIN Stefan Söllner
KONSTRUKTION VON ESCHERICHIA COLI PRODUKTIONSSTÄMMEN ZUR FERMENTATIVEN HERSTELLUNG VON SUCCINAT AUS GLYCERIN Von der Fakultät 4 (Energie-, Verfahrens- und Biotechnik) der Universität Stuttgart zur Erlangung der Würde eines Doktors der Naturwissenschaften (Dr. rer. nat.) genehmigte Abhandlung Vorgelegt von Stefan Söllner aus Schweinfurt Hauptberichter: Prof. Dr. rer. nat. Ralf Mattes Mitberichter: Prof. Dr.-Ing. Ralf Takors Tag der mündlichen Prüfung: 29.02.2012 Institut für Industrielle Genetik Universität Stuttgart 2012 Vielen Dank! … Herrn Prof. Dr. R. Mattes danke ich für die Überlassung des interessanten Themas, des Arbeitsplatzes, für gelegentliches Aufmuntern und für die Erstellung des Erstgutachtens dieser Arbeit. … Herrn Prof. Dr. R. Takors danke ich für die freundliche Übernahme des Zweitgutachtens und für spannende Diskussionen. … Herrn Dr. Josef Altenbuchner danke ich für die praktische Betreuung dieser Arbeit, für diverse Einladungen zu Grillfesten und ganz besonders für die intensive Durchsicht des Manuskriptes!!! … Herrn Dr. Martin Siemann-Herzberg danke ich für die motivierende, überschwängliche Begeisterung, die meine Ideen und Ergebnisse bei deren Besprechung jedesmal auslösten. … Herrn Prof. Dr. Reuss danke ich für die Initiierung des Projektes lange vor meiner Zeit. … Meinen Kollegen und Exkollegen danke ich für die gute Zusammenarbeit, das abwechslungsreiche Arbeitsklima sowie die vielen fachlichen und nichtfachlichen Gespräche, welche die Arbeit immer spannend gestalteten. Vor allem danke ich für das Verständnis für die von mir durchgeführten, absolut notwendigen, regelmäßigen Arbeitskontrollen. … Frau Dr. Anne Völker hat mir die Integration zu Beginn meines Aufenthaltes am IIG sehr erleichtert. Herzlichen Dank dafür! … Herrn Kambiz Morabbi Heravi danke ich recht herzlich für die Einladung ans National Institute of Genetic Engineering and Biotechnology in Teheran, Iran und für die internationale Freundschaft. -

Discovery of Novel Biomarkers for Atherosclerotic Aortic Aneurysm
www.nature.com/scientificreports There are amendments to this paper OPEN Discovery of novel biomarkers for atherosclerotic aortic aneurysm through proteomics-based assessment of disease progression Hiroaki Yagi1,6, Mitsuhiro Nishigori1,2,6, Yusuke Murakami2,6, Tsukasa Osaki1, Sayaka Muto2,4, Yutaka Iba3, Kenji Minatoya3, Yoshihiko Ikeda4, Hatsue Ishibashi-Ueda4, Takayuki Morisaki5, Hitoshi Ogino3, Hiroshi Tanaka3, Hiroaki Sasaki3, Hitoshi Matsuda3 & Naoto Minamino1,2* Since aortic aneurysms (AAs) are mostly asymptomatic, but they have a high mortality rate upon rupture, their detection and progression evaluation are clinically important issues. To discover diagnostic biomarkers for AA, we performed proteome analysis of aortic media from patients with thoracic atherosclerotic AA (TAAA), comparing protein levels between the aneurysm and normal tissue areas. After hierarchical clustering analysis of the proteome analysis data, tissue samples were classifed into three groups, regardless of morphological features. This classifcation was shown to refect disease progression stage identifed by pathological examination. This proteomics-based staging system enabled us to identify more signifcantly altered proteins than the morphological classifcation system. In subsequent data analysis, Niemann-Pick disease type C2 protein (NPC2) and insulin-like growth factor-binding protein 7 (IGFBP7) were selected as novel biomarker candidates for AA and were compared with the previously reported biomarker, thrombospondin 1 (THBS1). Blood concentrations of NPC2 and IGFBP7 were signifcantly increased, while THBS1 levels were decreased in TAAA and abdominal atherosclerotic AA patients. Receiver operating characteristic analysis of AA patients and healthy controls showed that NPC2 and IGFBP7 have higher specifcity and sensitivity than THBS1. Thus, NPC2 and IGFBP7 are promising biomarkers for the detection and progression evaluation of AA. -

A Systems Chemoproteomic Analysis of Acyl-Coa/Protein Interaction Networks
bioRxiv preprint doi: https://doi.org/10.1101/665281; this version posted July 18, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. A systems chemoproteomic analysis of acyl-CoA/protein interaction networks Michaella J. Levy,1# David C. Montgomery,2# Mihaela E. Sardiu,1 Sarah E. Bergholtz,2 Kellie D. Nance, 2 Jose Montano,2 Abigail L. Thorpe,2 Stephen D. Fox,3 Qishan Lin,4 Thorkell Andresson,3 Laurence Florens,1 Michael P. Washburn,1,5 and Jordan L. Meier2* 1. Stowers Institute for Medical Research, Kansas City, MO, 64110, USA 2. Chemical Biology Laboratory, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Frederick, MD, 21702, USA. 3. Laboratory of Proteomics and Analytical Technologies, Leidos, Inc, Frederick National Laboratory for Cancer Research, Frederick, Maryland 21702, USA 4. RNA Epitranscriptomics & Proteomics Resource, University of Albany, 1400 Washington Ave, Albany, NY 12222 5. Department of Pathology and Laboratory Medicine, University of Kansas Medical Center, Kansas City, KS, 66160, USA # M.J.L. and D.C.M. contributed equally to this work. * Lead contact to whom correspondence should be addressed. Email: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/665281; this version posted July 18, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Summary 2 Acyl-CoA/protein interactions are required for many functions essential to life including membrane synthesis, 3 oxidative metabolism, and macromolecular acetylation. -

Proteomics of Lipid Accumulation and DGAT Inhibition in Hepg2 Liver Carcinoma Cells
Proteomics of lipid accumulation and DGAT inhibition in HepG2 liver carcinoma cells. By Bhumika Bhatt-Wessel A thesis submitted to Victoria University of Wellington in fulfilment of the requirement for the degree of Doctor of Philosophy In Cell and Molecular Biology. Victoria University of Wellington 2017. i ABSTRACT Non-alcoholic fatty liver disease (NAFLD) is a manifestation of the metabolic syndrome in the liver. It is marked by hepatocyte accumulation of triacylglycerol (TAG) rich lipid droplets. In some patients, the disease progresses to non-alcoholic steatohepatitis (NASH), characterized by cellular damage, inflammation and fibrosis. In some cases, cirrhosis and liver failure may occur. However, the pathogenesis of NAFLD is still unclear. The present project is based on the hypothesis that hepatocytes are equipped with mechanisms that allow them to manage lipid accumulation to a certain extent. Continued or increased lipid accumulation beyond this triggers molecular mechanisms such as oxidative stress, lipid peroxidation and cell death that aggravate the condition and cause disease progression. The aim of this project is to study the effects of lipid accumulation on the cells using proteomics approach to identify proteins involved in the disease progression. A cell culture model was used in the study. HepG2 cells, a human liver carcinoma cell line, were treated with a mixture of fatty acids (FA) to induce lipid accumulation. The lipid accumulation in HepG2 cells was measured with Oil red O assay and the effect of lipid accumulation on the proliferation of the cells was measured using an MTT cell proliferation assay. HepG2 cells treated with 1 mM FA mixture for 6 hours induced lipid accumulation 1.4 times of control with 90% of cell proliferation capacity of the control cells. -

Supplemental Table 1. List of Candidate Gene Filters Used in the Analysis of Exome Sequencing. MYOPATHY NEUROPATHY MND ABHD5
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) J Neurol Neurosurg Psychiatry Supplemental table 1. List of candidate gene filters used in the analysis of exome sequencing. MYOPATHY NEUROPATHY MND ABHD5 AAAS AAAS ACADL AARS1 AARS1 ACADM ABCA1 AGT ACADS ABCD1 ALAD ACADVL ABHD12 ALS2 ACTA1 ADCY6 ANG ADSSL1 AFG3L2 APEX1 AGL AIFM1 APOE AGPAT2 AMACR AR AGRN ANG ASAH1 AIRE AP1S1 ATM ALDOA APOA1 ATP7A ALG14 APTX ATXN2 ALG2 ARHGEF10 ATXN3 ALG3 ARL6IP1 B4GALT6 ANKRD2 ARSA BCL11B ANO5 ASAH1 BCL6 ASCC1 ATL1 BICD2 ATGL ATL3 BSCL2 ATP2A1 ATM C19orf12 ATRN ATXN1 C9orf72 B3GALNT2 ATXN10 CCS B3GNT2 ATXN2 CDH13 BAG3 ATXN3 CDH22 BIN1 ATXN7 CHCHD10 BSCL2 B2M CHMP2B BVES B4GALNT1 CNTF CACNA1S BAG3 CNTN4 CAPN3 BCKDHB CNTN6 CASQ1 BSCL2 CRIM1 CAV1 C12orf65 CRYM CAV3 C9orf72 CSNK1G3 CAVIN1 CLP1 CST3 CCDC78 CNTNAP1 CUL4B CDKN1C COX10 CYP2D6 CFL2 COX6A1 DAO Grunseich C, et al. J Neurol Neurosurg Psychiatry 2021;0:1–11. doi: 10.1136/jnnp-2020-325437 BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) J Neurol Neurosurg Psychiatry CHAT CPOX DCAF15 CHCHD10 CRYAB DCTN1 CHD7 CTDP1 DIAPH3 CHKB CTSA DISC1 CHN1 CYP27A1 DNAJB2 CHRM3 DARS2 DOC2B CHRNA1 DDHD1 DPP6 CHRNB1 DGUOK DYNC1H1 CHRND DHH EFEMP1 CHRNE DHTKD1 ELP3 CIDEC DMD EPHA4 CLCN1 DNAJB2 EWSR1 CLN3 DNAJC3 EXOSC3 CNBP DNM2 FBLN5 CNTN1 DYNC1H1 FBXO38 COA3 EGR2 FEZF2 COL12A1 EMD FGGY COL13A1 ERCC6 FIG4 COL6A ERCC8 FUS COL6A1 FAH GARS1 COL6A2 FAM126A GBE1 COL6A3 FBLN5 GMPPA COL9A3 FGD4 GRB14 COLQ FGF14 GRN COX10 FIG4 HEXA COX15 FLNC HFE CPT2 FLRT1 HINT1 CRAT FLVCR1 HSPB1 CRPPA FMR1 HSPB3 CRYAB FUS HSPB8 CTNS FXN IGHMBP2 DAG1 GALC ITPR2 DECR1 GAN KDR DES GARS1 KIFAP3 DGUOK GBA2 KLHL9 DIH1 GBE1 LAMA2 DMD GDAP1 LAS1L DMPK GJB1 LIF DNAJB6 GJB3 LIPC DNAJC19 GLA LOX Grunseich C, et al. -

Fundamental Differences in Cell Cycle Deregulation in Human Papillomavirus–Positive and Human Papillomavirus–Negative Head/Neck and Cervical Cancers
Research Article Fundamental Differences in Cell Cycle Deregulation in Human Papillomavirus–Positive and Human Papillomavirus–Negative Head/Neck and Cervical Cancers Dohun Pyeon,1,2 Michael A. Newton,3 Paul F. Lambert,1 Johan A. den Boon,1,2 Srikumar Sengupta,1,2 Carmen J. Marsit,6 Craig D. Woodworth,7 Joseph P. Connor,4 Thomas H. Haugen,8 Elaine M. Smith,9 Karl T. Kelsey,6 Lubomir P. Turek,8 and Paul Ahlquist1,2,5 1McArdle Laboratory for Cancer Research; 2Institute for Molecular Virology; Departments of 3Statistics and of Biostatistics and Medical Informatics and 4Obstetrics and Gynecology; 5Howard Hughes Medical Institute, University of Wisconsin-Madison, Madison, Wisconsin; 6Departments of Genetics and Complex Diseases, Harvard School of Public Health, Boston, Massachusetts; 7Department of Biology, Clarkson University, Potsdam, New York; 8Department of Pathology, Veterans Affairs Medical Center; and 9Department of Epidemiology, University of Iowa, Iowa City, Iowa Abstract lower female reproductive tract, penis, and anus (1). In particular, Human papillomaviruses (HPV) are associated with nearly all high-risk HPVs are associated with nearly all cervical cancers, a cervical cancers, 20% to 30% of head and neck cancers (HNC), leading cause of cancer death in women worldwide despite the and other cancers. Because HNCs also arise in HPV-negative effectiveness in developed countries of screening for early patients, this type of cancer provides unique opportunities to detection of precancerous lesions (1). Prophylactic HPV vaccines define similarities and differences of HPV-positive versus HPV- should eventually reduce infections by the most prevalent high-risk HPVs, but do not cover all high-risk HPVs.