RNA-Seq Reveals Altered Gene Expression Levels in Proximal
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

SLC44A1 Transport of Choline and Ethanolamine in Disease
SLC44A1 Transport of Choline and Ethanolamine in Disease by Adrian Taylor A Thesis presented to The University of Guelph In partial fulfilment of requirements for the degree of Doctor of Philosophy in Human Health and Nutritional Sciences Guelph, Ontario, Canada © Adrian Taylor, April, 2019 ABSTRACT SLC44A1 TRANSPORT OF CHOLINE AND ETHANOLAMINE IN DISEASE Adrian Taylor Advisor(s): University of Guelph, 2019 Marica Bakovic Choline and ethanolamine are important molecules required for the de novo synthesis of phosphatidylcholine (PC) and phosphatidylethanolamine (PE) via the Kennedy pathway. Additionally, these two molecules are vital for maintaining both muscular and neurological function. The goal of this thesis was to gain insight into PC and PE metabolism with the use of unique metabolic disturbances ranging from obesity and genetic mutations in neurodegenerative disease. Firstly, the protective effects of choline supplementation on muscular function were investigated within the Pcyt2+/- mouse model. In Pcyt2+/- mice, substrate flow through the CDP-ethanolamine branch of the Kennedy pathway was diminished resulting in triacylglycerol (TAG) accumulation and obesity. Supplemental choline improved muscle function by altering the expression of genes devoted to reducing TAG synthesis and restoring energy homeostasis. With this new insight about the role of choline in regulating metabolism, the cellular uptake mechanism of choline was then analyzed. Skin fibroblasts from two patients with homozygous mutations in the SLC44A1 gene suffering from Neurodegeneration with Brain Iron Accumulation (NBIA) were utilized. In these fibroblasts, SLC44A1 expression and choline uptake were drastically diminished. Moreover, PC levels were unaffected while PE levels were diminished relative to control, an indication of perturbed phospholipid homeostasis. -

Upregulation of Peroxisome Proliferator-Activated Receptor-Α And
Upregulation of peroxisome proliferator-activated receptor-α and the lipid metabolism pathway promotes carcinogenesis of ampullary cancer Chih-Yang Wang, Ying-Jui Chao, Yi-Ling Chen, Tzu-Wen Wang, Nam Nhut Phan, Hui-Ping Hsu, Yan-Shen Shan, Ming-Derg Lai 1 Supplementary Table 1. Demographics and clinical outcomes of five patients with ampullary cancer Time of Tumor Time to Age Differentia survival/ Sex Staging size Morphology Recurrence recurrence Condition (years) tion expired (cm) (months) (months) T2N0, 51 F 211 Polypoid Unknown No -- Survived 193 stage Ib T2N0, 2.41.5 58 F Mixed Good Yes 14 Expired 17 stage Ib 0.6 T3N0, 4.53.5 68 M Polypoid Good No -- Survived 162 stage IIA 1.2 T3N0, 66 M 110.8 Ulcerative Good Yes 64 Expired 227 stage IIA T3N0, 60 M 21.81 Mixed Moderate Yes 5.6 Expired 16.7 stage IIA 2 Supplementary Table 2. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of an ampullary cancer microarray using the Database for Annotation, Visualization and Integrated Discovery (DAVID). This table contains only pathways with p values that ranged 0.0001~0.05. KEGG Pathway p value Genes Pentose and 1.50E-04 UGT1A6, CRYL1, UGT1A8, AKR1B1, UGT2B11, UGT2A3, glucuronate UGT2B10, UGT2B7, XYLB interconversions Drug metabolism 1.63E-04 CYP3A4, XDH, UGT1A6, CYP3A5, CES2, CYP3A7, UGT1A8, NAT2, UGT2B11, DPYD, UGT2A3, UGT2B10, UGT2B7 Maturity-onset 2.43E-04 HNF1A, HNF4A, SLC2A2, PKLR, NEUROD1, HNF4G, diabetes of the PDX1, NR5A2, NKX2-2 young Starch and sucrose 6.03E-04 GBA3, UGT1A6, G6PC, UGT1A8, ENPP3, MGAM, SI, metabolism -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Interplay Between Metformin and Serotonin Transport in the Gastrointestinal Tract: a Novel Mechanism for the Intestinal Absorption and Adverse Effects of Metformin
INTERPLAY BETWEEN METFORMIN AND SEROTONIN TRANSPORT IN THE GASTROINTESTINAL TRACT: A NOVEL MECHANISM FOR THE INTESTINAL ABSORPTION AND ADVERSE EFFECTS OF METFORMIN Tianxiang Han A dissertation submitted to the faculty of the University of North Carolina at Chapel Hill in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Eshelman School of Pharmacy. Chapel Hill 2013 Approved By: Dhiren R. Thakker, Ph.D. Michael Jay, Ph.D. Kim L. R. Brouwer, Pharm.D., Ph.D. Joseph W. Polli, Ph.D. Xiao Xiao, Ph.D. © 2013 Tianxiang Han ALL RIGHTS RESERVED ii ABSTRACT TIANXIANG HAN: Interplay between Metformin and Serotonin Transport in the Gastrointestinal Tract: A Novel Mechanism for the Intestinal Absorption and Adverse Effects of Metformin (Under the direction of Dhiren R. Thakker, Ph.D.) Metformin is a widely prescribed drug for Type II diabetes mellitus. Previous studies have shown that this highly hydrophilic and charged compound traverses predominantly paracellularly across the Caco-2 cell monolayer, a well-established model for human intestinal epithelium. However, oral bioavailability of metformin is significantly higher than that of the paracellular probe, mannitol (~60% vs ~16%). Based on these observations, the Thakker laboratory proposed a “sponge” hypothesis (Proctor et al., 2008) which states that the functional synergy between apical (AP) transporters and paracellular transport enhances the intestinal absorption of metformin. This dissertation work aims to identify AP uptake transporters of metformin, determine their polarized localization, and elucidate their roles in the intestinal absorption and adverse effects of metformin. Chemical inhibition and transporter-knockdown studies revealed that four transporters, namely, organic cation transporter 1 (OCT1), plasma membrane monoamine transporter (PMAT), serotonin reuptake transporter (SERT) and choline high-affinity transporter (CHT) contribute to AP uptake of metformin in Caco-2 cells. -
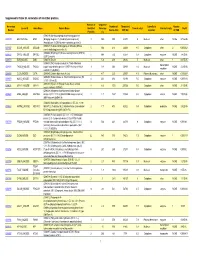
Supporting Table S3 for PDF Maker
Supplemental Table S3. Annotation of identified proteins. Number of Sequence Accession Number of Theoretical Subcellular Number Locus ID Gene Name Protein Name Identified Coverage Theoretical pI Protein Family NSAF Number Amino Acid MW (Da) Location of TMD Peptides (%) (O08539) Myc box-dependent-interacting protein 1 O08539 BIN1_MOUSE BIN1 (Bridging integrator 1) (Amphiphysin-like protein) 3 10.5 588 64470 5 Nucleus other NONE 5.73E-05 (Amphiphysin II) (SH3-domain-containing protein 9) (O08547) Vesicle-trafficking protein SEC22b (SEC22 O08547 SC22B_MOUSE SEC22B 5 30.4 214 24609 8.5 Cytoplasm other 2 0.000262 vesicle-trafficking protein-like 1) (O08553) Dihydropyrimidinase-related protein 2 (DRP-2) O08553 DPYL2_MOUSE DPYSL2 5 19.4 572 62278 6.4 Cytoplasm enzyme NONE 7.85E-05 (ULIP 2 protein) O08579 EMD_MOUSE EMD (O08579) Emerin 1 5.8 259 29436 5 Nucleus other 1 8.67E-05 (O08583) THO complex subunit 4 (Tho4) (RNA and transcription O08583 THOC4_MOUSE THOC4 export factor-binding protein 1) (REF1-I) (Ally of AML-1 1 9.8 254 26809 11.2 Nucleus NONE 2.21E-05 regulator and LEF-1) (Aly/REF) O08585 CLCA_MOUSE CLTA (O08585) Clathrin light chain A (Lca) 2 4.7 235 25557 4.5 Plasma Membrane other NONE 0.000287 (O08600) Endonuclease G, mitochondrial precursor (EC O08600 NUCG_MOUSE ENDOG 4 23.1 294 32191 9.5 Cytoplasm enzyme NONE 0.000134 3.1.30.-) (Endo G) (O08638) Myosin-11 (Myosin heavy chain, smooth O08638 MYH11_MOUSE MYH11 8 6.6 1972 227026 5.5 Cytoplasm other NONE 3.13E-05 muscle isoform) (SMMHC) (O08648) Mitogen-activated protein kinase kinase -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

KONSTRUKTION VON ESCHERICHIA COLI PRODUKTIONSSTÄMMEN ZUR FERMENTATIVEN HERSTELLUNG VON SUCCINAT AUS GLYCERIN Stefan Söllner
KONSTRUKTION VON ESCHERICHIA COLI PRODUKTIONSSTÄMMEN ZUR FERMENTATIVEN HERSTELLUNG VON SUCCINAT AUS GLYCERIN Von der Fakultät 4 (Energie-, Verfahrens- und Biotechnik) der Universität Stuttgart zur Erlangung der Würde eines Doktors der Naturwissenschaften (Dr. rer. nat.) genehmigte Abhandlung Vorgelegt von Stefan Söllner aus Schweinfurt Hauptberichter: Prof. Dr. rer. nat. Ralf Mattes Mitberichter: Prof. Dr.-Ing. Ralf Takors Tag der mündlichen Prüfung: 29.02.2012 Institut für Industrielle Genetik Universität Stuttgart 2012 Vielen Dank! … Herrn Prof. Dr. R. Mattes danke ich für die Überlassung des interessanten Themas, des Arbeitsplatzes, für gelegentliches Aufmuntern und für die Erstellung des Erstgutachtens dieser Arbeit. … Herrn Prof. Dr. R. Takors danke ich für die freundliche Übernahme des Zweitgutachtens und für spannende Diskussionen. … Herrn Dr. Josef Altenbuchner danke ich für die praktische Betreuung dieser Arbeit, für diverse Einladungen zu Grillfesten und ganz besonders für die intensive Durchsicht des Manuskriptes!!! … Herrn Dr. Martin Siemann-Herzberg danke ich für die motivierende, überschwängliche Begeisterung, die meine Ideen und Ergebnisse bei deren Besprechung jedesmal auslösten. … Herrn Prof. Dr. Reuss danke ich für die Initiierung des Projektes lange vor meiner Zeit. … Meinen Kollegen und Exkollegen danke ich für die gute Zusammenarbeit, das abwechslungsreiche Arbeitsklima sowie die vielen fachlichen und nichtfachlichen Gespräche, welche die Arbeit immer spannend gestalteten. Vor allem danke ich für das Verständnis für die von mir durchgeführten, absolut notwendigen, regelmäßigen Arbeitskontrollen. … Frau Dr. Anne Völker hat mir die Integration zu Beginn meines Aufenthaltes am IIG sehr erleichtert. Herzlichen Dank dafür! … Herrn Kambiz Morabbi Heravi danke ich recht herzlich für die Einladung ans National Institute of Genetic Engineering and Biotechnology in Teheran, Iran und für die internationale Freundschaft. -

SLC22A5 Mutations in a Patient with Systemic Primary Carnitine Deficiency: the First Korean Case Confirmed by Biochemical and Molecular Investigation
Available online at www.annclinlabsci.org 424 Annals of Clinical & Laboratory Science, vol. 42, no. 4, 2012 SLC22A5 Mutations in a Patient with Systemic Primary Carnitine Deficiency: The First Korean Case Confirmed by Biochemical and Molecular Investigation Young Ahn Yoon1, Dong Hwan Lee2, Chang-Seok Ki1, Soo-Youn Lee1, Jong-Won Kim1, Yong-Wha Lee3, and Hyung-Doo Park1 1Department of Laboratory Medicine and Genetics, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul; 2Department of Pediatrics, Soonchunhyang University College of Medicine, Seoul, Korea; 3Depart- ment of Laboratory Medicine and Genetics, Soonchunhyang University Bucheon Hospital, Soonchunhyang University College of Medicine, Bucheon, Korea. Abstract. Systemic primary carnitine deficiency (CDSP) is a rare autosomal recessive disorder that pres- ents episodic periods of hypoketotic hypoglycemia. The main symptoms of CDSP are skeletal and cardiac myopathy. CDSP is caused by a defect in plasma membrane uptake of carnitine, ultimately caused by the SLC22A5 gene. We report the case of a Korean patient with CDSP. He had an abnormal free carnitine level of 5.56 µmol/L (reference range, RR 10.4~87.1 µmol/L) and a palmitoylcarnitine level of 0.27 µmol/L (RR 0.5~9.7 µmol/L) in a newborn screening test. The patient showed an ammonia level of 129.4 ug/dL (RR, 25~65 ug/dL), a lactate level of 4.5 mmol/L (RR, 0.5-2.2 mmol/L), and a free carnitine level of 10.3 µmol/L (RR, 36-74 µmol/L) in blood. After PCR-sequencing analysis of the SLC22A5 gene, the patient was found to be a compound heterozygote for c.506G>A (p.R169Q) and c.1400C>G (p.S467C) muta- tions. -

Discovery of Novel Biomarkers for Atherosclerotic Aortic Aneurysm
www.nature.com/scientificreports There are amendments to this paper OPEN Discovery of novel biomarkers for atherosclerotic aortic aneurysm through proteomics-based assessment of disease progression Hiroaki Yagi1,6, Mitsuhiro Nishigori1,2,6, Yusuke Murakami2,6, Tsukasa Osaki1, Sayaka Muto2,4, Yutaka Iba3, Kenji Minatoya3, Yoshihiko Ikeda4, Hatsue Ishibashi-Ueda4, Takayuki Morisaki5, Hitoshi Ogino3, Hiroshi Tanaka3, Hiroaki Sasaki3, Hitoshi Matsuda3 & Naoto Minamino1,2* Since aortic aneurysms (AAs) are mostly asymptomatic, but they have a high mortality rate upon rupture, their detection and progression evaluation are clinically important issues. To discover diagnostic biomarkers for AA, we performed proteome analysis of aortic media from patients with thoracic atherosclerotic AA (TAAA), comparing protein levels between the aneurysm and normal tissue areas. After hierarchical clustering analysis of the proteome analysis data, tissue samples were classifed into three groups, regardless of morphological features. This classifcation was shown to refect disease progression stage identifed by pathological examination. This proteomics-based staging system enabled us to identify more signifcantly altered proteins than the morphological classifcation system. In subsequent data analysis, Niemann-Pick disease type C2 protein (NPC2) and insulin-like growth factor-binding protein 7 (IGFBP7) were selected as novel biomarker candidates for AA and were compared with the previously reported biomarker, thrombospondin 1 (THBS1). Blood concentrations of NPC2 and IGFBP7 were signifcantly increased, while THBS1 levels were decreased in TAAA and abdominal atherosclerotic AA patients. Receiver operating characteristic analysis of AA patients and healthy controls showed that NPC2 and IGFBP7 have higher specifcity and sensitivity than THBS1. Thus, NPC2 and IGFBP7 are promising biomarkers for the detection and progression evaluation of AA. -

The Metabolomic Paradigm of Pharmacogenomics in Complex
ics: O om pe ol n b A a c t c e e M s s Cacabelos, Metabolomics 2012, 2:5 Metabolomics: Open Access DOI: 10.4172/2153-0769.1000e119 ISSN: 2153-0769 Editorial Open Access The Metabolomic Paradigm of Pharmacogenomics in Complex Disorders Ramón Cacabelos1,2* 1EuroEspes Biomedical Research Center, Institute for CNS Disorders and Genomic Medicine, EuroEspes Chair of Biotechnology and Genomics, 15165-Bergondo, Corunna, Spain 2President, World Association of Genomic Medicine, Spain Metabolomics represents the networking organization of multiple pathogenic genes usually converge in metabolomic networks leading biochemical pathways leading to a physiological function in living to specific pathogenic cascades responsible for disease phenotypes. organisms. The frontier between health and disease is likely to be the The genomics of the mechanism of action of drugs has so far been result of a fine-tuning equilibrium or disequilibrium, respectively, neglected by the scientific community, and consequently less than 5% between the genomic-transcriptomic-proteomic-metabolomic cascade of FDA-approved drugs, with a pharmacologically-defined mechanism and environmental factors and/or epigenetic phenomena. In recent of action, have been studied in order to evaluate whether mutations times, diverse metabolomic studies have emerged in medical science in the genes encoding receptors or enzymes may affect efficacy and to explain physiological and pathogenic events in several disciplines, safety issues. Pleiotropic genes, involved in multiple pathogenic events, -

Analysis of OAT, OCT, OCTN, and Other Family Members Reveals 8
bioRxiv preprint doi: https://doi.org/10.1101/2019.12.23.887299; this version posted December 26, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Reclassification of SLC22 Transporters: Analysis of OAT, OCT, OCTN, and other Family Members Reveals 8 Functional Subgroups Darcy Engelhart1, Jeffry C. Granados2, Da Shi3, Milton Saier Jr.4, Michael Baker6, Ruben Abagyan3, Sanjay K. Nigam5,6 1Department of Biology, University of California San Diego, La Jolla 92093 2Department of Bioengineering, University of California San Diego, La Jolla 92093 3School of Pharmacy and Pharmaceutical Sciences, University of California San Diego, La Jolla 92093 4Department of Molecular Biology, Division of Biological Sciences, University of California at San Diego, San Diego, CA, USA 5Department of Pediatrics, University of California San Diego, La Jolla 92093 6Department of Medicine, University of California San Diego, La Jolla 92093 *To whom correspondence should be addressed: [email protected] Running title: Functional subgroups for SLC22 1 bioRxiv preprint doi: https://doi.org/10.1101/2019.12.23.887299; this version posted December 26, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Abstract Among transporters, the SLC22 family is emerging as a central hub of endogenous physiology. -

Disease-Induced Modulation of Drug Transporters at the Blood–Brain Barrier Level
International Journal of Molecular Sciences Review Disease-Induced Modulation of Drug Transporters at the Blood–Brain Barrier Level Sweilem B. Al Rihani 1 , Lucy I. Darakjian 1, Malavika Deodhar 1 , Pamela Dow 1 , Jacques Turgeon 1,2 and Veronique Michaud 1,2,* 1 Tabula Rasa HealthCare, Precision Pharmacotherapy Research and Development Institute, Orlando, FL 32827, USA; [email protected] (S.B.A.R.); [email protected] (L.I.D.); [email protected] (M.D.); [email protected] (P.D.); [email protected] (J.T.) 2 Faculty of Pharmacy, Université de Montréal, Montreal, QC H3C 3J7, Canada * Correspondence: [email protected]; Tel.: +1-856-938-8697 Abstract: The blood–brain barrier (BBB) is a highly selective and restrictive semipermeable network of cells and blood vessel constituents. All components of the neurovascular unit give to the BBB its crucial and protective function, i.e., to regulate homeostasis in the central nervous system (CNS) by removing substances from the endothelial compartment and supplying the brain with nutrients and other endogenous compounds. Many transporters have been identified that play a role in maintaining BBB integrity and homeostasis. As such, the restrictive nature of the BBB provides an obstacle for drug delivery to the CNS. Nevertheless, according to their physicochemical or pharmacological properties, drugs may reach the CNS by passive diffusion or be subjected to putative influx and/or efflux through BBB membrane transporters, allowing or limiting their distribution to the CNS. Drug transporters functionally expressed on various compartments of the BBB involve numerous proteins from either the ATP-binding cassette (ABC) or the solute carrier (SLC) superfamilies.