Distribution and Evolution of the Mobile Vma-1B Intein
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Growth of Sedimentary Bathyarchaeota on Lignin As an Energy Source
Growth of sedimentary Bathyarchaeota on lignin as an energy source Tiantian Yua,b,1, Weichao Wuc,d,1, Wenyue Lianga,b, Mark Alexander Levere, Kai-Uwe Hinrichsc,d, and Fengping Wanga,b,2 aState Key Laboratory of Microbial Metabolism, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, 200240 Shanghai, China; bState Key Laboratory of Ocean Engineering, Shanghai Jiao Tong University, 200240 Shanghai, China; cOrganic Geochemistry Group, MARUM-Center for Marine Environmental Sciences, University of Bremen, 28359 Bremen, Germany; dDepartment of Geosciences, University of Bremen, 28359 Bremen, Germany; and eInstitute of Biogeochemistry and Pollutant Dynamics, Department of Environmental Systems Science, Swiss Federal Institute of Technology Zurich, 8092 Zurich, Switzerland Edited by Edward F. DeLong, University of Hawaii at Manoa, Honolulu, HI, and approved April 16, 2018 (received for review October 30, 2017) Members of the archaeal phylum Bathyarchaeota are among the other hand, acetogenesis from H2/CO2 has been proposed for most abundant microorganisms on Earth. Although versatile met- some lineages of Bathyarchaeota (3). The reductive acetyl-CoA abolic capabilities such as acetogenesis, methanogenesis, and fer- pathway for carbon fixation has been identified in most of the mentation have been suggested for bathyarchaeotal members, no obtained bathyarchaeotal genomes (3, 12, 14). Among these, direct confirmation of these metabolic functions has been achieved members of the bathyarchaeotal subgroups Bathy-3 and Bathy- through growth of Bathyarchaeota in the laboratory. Here we dem- 8 have been suggested to be capable of methanogenesis (12). All onstrate, on the basis of gene-copy numbers and probing of ar- the above studies indicate great versatility in the metabolic po- chaeal lipids, the growth of Bathyarchaeota subgroup Bathy-8 in tentials of Bathyarchaeota, with some lineages possibly being enrichments of estuarine sediments with the biopolymer lignin. -

Phylogenetics of Archaeal Lipids Amy Kelly 9/27/2006 Outline
Phylogenetics of Archaeal Lipids Amy Kelly 9/27/2006 Outline • Phlogenetics of Archaea • Phlogenetics of archaeal lipids • Papers Phyla • Two? main phyla – Euryarchaeota • Methanogens • Extreme halophiles • Extreme thermophiles • Sulfate-reducing – Crenarchaeota • Extreme thermophiles – Korarchaeota? • Hyperthermophiles • indicated only by environmental DNA sequences – Nanoarchaeum? • N. equitans a fast evolving euryarchaeal lineage, not novel, early diverging archaeal phylum – Ancient archael group? • In deepest brances of Crenarchaea? Euryarchaea? Archaeal Lipids • Methanogens – Di- and tetra-ethers of glycerol and isoprenoid alcohols – Core mostly archaeol or caldarchaeol – Core sometimes sn-2- or Images removed due to sn-3-hydroxyarchaeol or copyright considerations. macrocyclic archaeol –PMI • Halophiles – Similar to methanogens – Exclusively synthesize bacterioruberin • Marine Crenarchaea Depositional Archaeal Lipids Biological Origin Environment Crocetane methanotrophs? methane seeps? methanogens, PMI (2,6,10,15,19-pentamethylicosane) methanotrophs hypersaline, anoxic Squalane hypersaline? C31-C40 head-to-head isoprenoids Smit & Mushegian • “Lost” enzymes of MVA pathway must exist – Phosphomevalonate kinase (PMK) – Diphosphomevalonate decarboxylase – Isopentenyl diphosphate isomerase (IPPI) Kaneda et al. 2001 Rohdich et al. 2001 Boucher et al. • Isoprenoid biosynthesis of archaea evolved through a combination of processes – Co-option of ancestral enzymes – Modification of enzymatic specificity – Orthologous and non-orthologous gene -

Characteristics and Metabolic Patterns of Soil Methanogenic Archaea Communities in the High Latitude Natural Wetlands of China
Characteristics and Metabolic Patterns of Soil Methanogenic Archaea Communities in the High Latitude Natural Wetlands of China Di Wu Northeast Forestry University Caihong Zhao Northeast Forestry University Hui Bai Forestry Science Research Institute of Heilongjiang Province Fujuan Feng Northeast Forestry University Xin Sui Heilongjiang University Guangyu Sun ( [email protected] ) Northeast Forestry University Research article Keywords: Wetlands, Methanogens, Community diversity, Indicator species, Methanogenic metabolic patterns Posted Date: August 12th, 2020 DOI: https://doi.org/10.21203/rs.3.rs-54821/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/18 Abstract Background: Soil methanogenic microorganisms are one of the primary methane-producing microbes in wetlands. However, we still poorly understand the community characteristic and metabolic patterns of these microorganisms according to vegetation type and seasonal changes. Therefore, to better elucidate the effects of the vegetation type and seasonal factors on the methanogenic community structure and metabolic patterns, we detected the characteristics of the soil methanogenic mcrA gene from three types of natural wetlands in different seasons in the Xiaoxing'an Mountain region, China. Result: The results indicated that the distribution of Methanobacteriaceae (hydrogenotrophic methanogens) was higher in winter, while Methanosarcinaceae and Methanosaetaceae accounted for a higher proportion in summer. Hydrogenotrophic methanogenesis was the dominant trophic pattern in each wetland. The results of principal coordinate analysis and cluster analysis showed that the vegetation type considerably inuenced the methanogenic community composition. The methanogenic community structure in the Betula platyphylla – Larix gmelinii wetland was relatively different from the structure of the other two wetland types. -

2I52 Lichtarge Lab 2006
Pages 1–11 2i52 Evolutionary trace report by report maker February 25, 2010 4.3.3 DSSP 10 4.3.4 HSSP 10 4.3.5 LaTex 10 4.3.6 Muscle 10 4.3.7 Pymol 10 4.4 Note about ET Viewer 10 4.5 Citing this work 10 4.6 About report maker 11 4.7 Attachments 11 1 INTRODUCTION From the original Protein Data Bank entry (PDB id 2i52): Title: Crystal structure of protein pto0218 from picrophilus torridus, pfam duf372 Compound: Mol id: 1; molecule: hypothetical protein; chain: a, b, CONTENTS c, d, e, f; engineered: yes Organism, scientific name: Picrophilus Torridus; 1 Introduction 1 2i52 contains a single unique chain 2i52F (119 residues long) and its homologues 2i52A, 2i52D, 2i52C, 2i52E, and 2i52B. 2 Chain 2i52F 1 2.1 Q6L2J9 overview 1 2.2 Multiple sequence alignment for 2i52F 1 2.3 Residue ranking in 2i52F 1 2.4 Top ranking residues in 2i52F and their position on the structure 1 2 CHAIN 2I52F 2.4.1 Clustering of residues at 25% coverage. 1 2.4.2 Overlap with known functional surfaces at 2.1 Q6L2J9 overview 25% coverage. 2 From SwissProt, id Q6L2J9, 97% identical to 2i52F: Description: Hypothetical protein. 3 Notes on using trace results 9 Organism, scientific name: Picrophilus torridus. 3.1 Coverage 9 Taxonomy: Archaea; Euryarchaeota; Thermoplasmata; Thermoplas- 3.2 Known substitutions 9 matales; Picrophilaceae; Picrophilus. 3.3 Surface 9 3.4 Number of contacts 9 3.5 Annotation 9 3.6 Mutation suggestions 9 2.2 Multiple sequence alignment for 2i52F 4 Appendix 9 For the chain 2i52F, the alignment 2i52F.msf (attached) with 30 4.1 File formats 9 sequences was used. -

Proteome Cold-Shock Response in the Extremely Acidophilic Archaeon, Cuniculiplasma Divulgatum
microorganisms Article Proteome Cold-Shock Response in the Extremely Acidophilic Archaeon, Cuniculiplasma divulgatum Rafael Bargiela 1 , Karin Lanthaler 1,2, Colin M. Potter 1,2 , Manuel Ferrer 3 , Alexander F. Yakunin 1,2, Bela Paizs 1,2, Peter N. Golyshin 1,2 and Olga V. Golyshina 1,2,* 1 School of Natural Sciences, Bangor University, Deiniol Rd, Bangor LL57 2UW, UK; [email protected] (R.B.); [email protected] (K.L.); [email protected] (C.M.P.); [email protected] (A.F.Y.); [email protected] (B.P.); [email protected] (P.N.G.) 2 Centre for Environmental Biotechnology, Bangor University, Deiniol Rd, Bangor LL57 2UW, UK 3 Systems Biotechnology Group, Department of Applied Biocatalysis, CSIC—Institute of Catalysis, Marie Curie 2, 28049 Madrid, Spain; [email protected] * Correspondence: [email protected]; Tel.: +44-1248-388607; Fax: +44-1248-382569 Received: 27 April 2020; Accepted: 15 May 2020; Published: 19 May 2020 Abstract: The archaeon Cuniculiplasma divulgatum is ubiquitous in acidic environments with low-to-moderate temperatures. However, molecular mechanisms underlying its ability to thrive at lower temperatures remain unexplored. Using mass spectrometry (MS)-based proteomics, we analysed the effect of short-term (3 h) exposure to cold. The C. divulgatum genome encodes 2016 protein-coding genes, from which 819 proteins were identified in the cells grown under optimal conditions. In line with the peptidolytic lifestyle of C. divulgatum, its intracellular proteome revealed the abundance of proteases, ABC transporters and cytochrome C oxidase. From 747 quantifiable polypeptides, the levels of 582 proteins showed no change after the cold shock, whereas 104 proteins were upregulated suggesting that they might be contributing to cold adaptation. -

Insights Into Archaeal Evolution and Symbiosis from the Genomes of a Nanoarchaeon and Its Inferred Crenarchaeal Host from Obsidian Pool, Yellowstone National Park
University of Tennessee, Knoxville TRACE: Tennessee Research and Creative Exchange Microbiology Publications and Other Works Microbiology 4-22-2013 Insights into archaeal evolution and symbiosis from the genomes of a nanoarchaeon and its inferred crenarchaeal host from Obsidian Pool, Yellowstone National Park Mircea Podar University of Tennessee - Knoxville, [email protected] Kira S. Makarova National Institutes of Health David E. Graham University of Tennessee - Knoxville, [email protected] Yuri I. Wolf National Institutes of Health Eugene V. Koonin National Institutes of Health See next page for additional authors Follow this and additional works at: https://trace.tennessee.edu/utk_micrpubs Part of the Microbiology Commons Recommended Citation Biology Direct 2013, 8:9 doi:10.1186/1745-6150-8-9 This Article is brought to you for free and open access by the Microbiology at TRACE: Tennessee Research and Creative Exchange. It has been accepted for inclusion in Microbiology Publications and Other Works by an authorized administrator of TRACE: Tennessee Research and Creative Exchange. For more information, please contact [email protected]. Authors Mircea Podar, Kira S. Makarova, David E. Graham, Yuri I. Wolf, Eugene V. Koonin, and Anna-Louise Reysenbach This article is available at TRACE: Tennessee Research and Creative Exchange: https://trace.tennessee.edu/ utk_micrpubs/44 Podar et al. Biology Direct 2013, 8:9 http://www.biology-direct.com/content/8/1/9 RESEARCH Open Access Insights into archaeal evolution and symbiosis from the genomes of a nanoarchaeon and its inferred crenarchaeal host from Obsidian Pool, Yellowstone National Park Mircea Podar1,2*, Kira S Makarova3, David E Graham1,2, Yuri I Wolf3, Eugene V Koonin3 and Anna-Louise Reysenbach4 Abstract Background: A single cultured marine organism, Nanoarchaeum equitans, represents the Nanoarchaeota branch of symbiotic Archaea, with a highly reduced genome and unusual features such as multiple split genes. -

Picrophilus Oshimae and Picrophilus Tomdus Fam. Nov., Gen. Nov., Sp. Nov
INTERNATIONALJOURNAL OF SYSTEMATICBACTERIOLOGY, July 1996, p. 814-816 Vol. 46, No. 3 0020-77 13/96/$04.00+0 Copyright 0 1996, International Union of Microbiological Societies Picrophilus oshimae and Picrophilus tomdus fam. nov., gen. nov., sp. nov., Two Species of Hyperacidophilic, Thermophilic, Heterotrophic, Aerobic Archaea CHRISTA SCHLEPER, GABRIELA PUHLER, HANS- PETER KLENK, AND WOLFRAM ZILLIG* Max Plank Institut fur Biochemie, 0-82152 Martinsried, Germany We describe two species of hyperacidophilic, thermophilic, heterotrophic, aerobic archaea that were isolated from solfataric hydrothermal areas in Hokkaido, Japan. These organisms, Picrophilus oshimae and Picrophilus torridus, represent a novel genus and a novel family, the Picrophilaceae, in the kingdom Euryarchaeota and the order Thermoplasmales. Both of these bacteria are more acidophilic than the genus Thermoplasma since they are able to grow at about pH 0. The moderately thermophilic, hyperacidophilic, aerobic ar- which comprises acid-loving (i.e., hyperacidophilic) organisms. chaea (archaebacteria) (7) Picrophilus oshimae and Picrophilus Separation of these taxa is justified by their phylogenetic dis- rorridus, which have been described previously (4, 5), were tance, (9.5% difference in the 16s rRNA sequences of mem- isolated from moderately hot hydrothermal areas in solfataric bers of the Picrophilaceae and T. acidophilum), by the lack of fields in Hokkaido, Japan. One of the sources of isolation was immunochemical cross-reactions in Ouchterlony immunodif- a solfataric spring which had a temperature of 53°C and a pH fusion assays between the RNA polymerases of P. oshimae and of 2.2, and the other was a rather dry hot soil which had a pH T. acidophilum, which also do not occur between members of of <OS. -
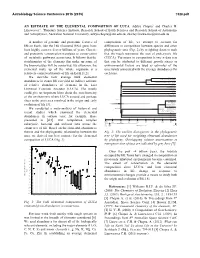
An Estimate of the Elemental Composition of Luca
Astrobiology Science Conference 2015 (2015) 7328.pdf AN ESTIMATE OF THE ELEMENTAL COMPOSITION OF LUCA. Aditya Chopra1 and Charles H. Lineweaver1, 1Planetary Science Institute, Research School of Earth Sciences and Research School of Astronomy and Astrophysics, Australian National University, [email protected], [email protected] A number of genomic and proteomic features of composition of life, we attempt to account for life on Earth, like the 16S ribosomal RNA gene, have differences in composition between species and other been highly conserved over billions of years. Genetic phylogenetic taxa (Fig. 2) by weighting datasets such and proteomic conservation translates to conservation that the result represents the root of prokaryotic life of metabolic pathways across taxa. It follows that the (LUCA). Variations in composition between data sets stoichiometry of the elements that make up some of that can be attributed to different growth stages or the biomolecules will be conserved. By extension, the environmental factors are used as estimates of the elemental make up of the whole organism is a uncertainty associated with the average abundances for relatively conserved feature of life on Earth [1,2]. each taxa. We describe how average bulk elemental Euryarchaeota 1a Methanococci, Methanobacteria, Methanopyri 7 Euryarchaeota 1b abundances in extant life can yield an indirect estimate 6 Thermoplasmata, Methanomicrobia, Halobacteria, Archaeoglobi Euryarchaeota 2 4 Thermococci of relative abundances of elements in the Last Crenarchaeota Sulfolobus, Thermoproteus ? Thaumarchaeota Universal Common Ancestor (LUCA). The results Cenarchaeum ? Korarchaeota ARMAN could give us important hints about the stoichiometry ? 2 Archaeal Richmond Mine Acidophilic Nanoorganisms Nanoarchaeota of the environment where LUCA existed and perhaps Archaea Terrabacteria clues to the processes involved in the origin and early Actinobacteria, Deinococcus-Thermus, 1 Cyanobacteria, Life Chloroflexi, 9 evolution of life [3]. -

Pan-Genome Analysis and Ancestral State Reconstruction Of
www.nature.com/scientificreports OPEN Pan‑genome analysis and ancestral state reconstruction of class halobacteria: probability of a new super‑order Sonam Gaba1,2, Abha Kumari2, Marnix Medema 3 & Rajeev Kaushik1* Halobacteria, a class of Euryarchaeota are extremely halophilic archaea that can adapt to a wide range of salt concentration generally from 10% NaCl to saturated salt concentration of 32% NaCl. It consists of the orders: Halobacteriales, Haloferaciales and Natriabales. Pan‑genome analysis of class Halobacteria was done to explore the core (300) and variable components (Softcore: 998, Cloud:36531, Shell:11784). The core component revealed genes of replication, transcription, translation and repair, whereas the variable component had a major portion of environmental information processing. The pan‑gene matrix was mapped onto the core‑gene tree to fnd the ancestral (44.8%) and derived genes (55.1%) of the Last Common Ancestor of Halobacteria. A High percentage of derived genes along with presence of transformation and conjugation genes indicate the occurrence of horizontal gene transfer during the evolution of Halobacteria. A Core and pan‑gene tree were also constructed to infer a phylogeny which implicated on the new super‑order comprising of Natrialbales and Halobacteriales. Halobacteria1,2 is a class of phylum Euryarchaeota3 consisting of extremely halophilic archaea found till date and contains three orders namely Halobacteriales4,5 Haloferacales5 and Natrialbales5. Tese microorganisms are able to dwell at wide range of salt concentration generally from 10% NaCl to saturated salt concentration of 32% NaCl6. Halobacteria, as the name suggests were once considered a part of a domain "Bacteria" but with the discovery of the third domain "Archaea" by Carl Woese et al.7, it became part of Archaea. -

Phylogeny and Taxonomy of Archaea: a Comparison of the Whole-Genome-Based Cvtree Approach with 16S Rrna Sequence Analysis
Life 2015, 5, 949-968; doi:10.3390/life5010949 OPEN ACCESS life ISSN 2075-1729 www.mdpi.com/journal/life Article Phylogeny and Taxonomy of Archaea: A Comparison of the Whole-Genome-Based CVTree Approach with 16S rRNA Sequence Analysis Guanghong Zuo 1, Zhao Xu 2 and Bailin Hao 1;* 1 T-Life Research Center and Department of Physics, Fudan University, 220 Handan Road, Shanghai 200433, China; E-Mail: [email protected] 2 Thermo Fisher Scientific, 200 Oyster Point Blvd, South San Francisco, CA 94080, USA; E-Mail: [email protected] * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +86-21-6565-2305. Academic Editors: Roger A. Garrett, Hans-Peter Klenk and Michael W. W. Adams Received: 9 December 2014 / Accepted: 9 March 2015 / Published: 17 March 2015 Abstract: A tripartite comparison of Archaea phylogeny and taxonomy at and above the rank order is reported: (1) the whole-genome-based and alignment-free CVTree using 179 genomes; (2) the 16S rRNA analysis exemplified by the All-Species Living Tree with 366 archaeal sequences; and (3) the Second Edition of Bergey’s Manual of Systematic Bacteriology complemented by some current literature. A high degree of agreement is reached at these ranks. From the newly proposed archaeal phyla, Korarchaeota, Thaumarchaeota, Nanoarchaeota and Aigarchaeota, to the recent suggestion to divide the class Halobacteria into three orders, all gain substantial support from CVTree. In addition, the CVTree helped to determine the taxonomic position of some newly sequenced genomes without proper lineage information. A few discrepancies between the CVTree and the 16S rRNA approaches call for further investigation. -

Article Mode, Which Adds to a Better and Living Conditions of Microorganisms in the Atmosphere Understanding of the Overall Atmospheric Microbiome
Biogeosciences, 15, 4205–4214, 2018 https://doi.org/10.5194/bg-15-4205-2018 © Author(s) 2018. This work is distributed under the Creative Commons Attribution 4.0 License. Community composition and seasonal changes of archaea in coarse and fine air particulate matter Jörn Wehking1,2, Daniel A. Pickersgill1,2, Robert M. Bowers3,4, David Teschner1,2, Ulrich Pöschl2, Janine Fröhlich-Nowoisky2, and Viviane R. Després1,2 1Institute of Molecular Physiology, Johannes Gutenberg University, Johannes-von-Müller-Weg 6, 55128 Mainz, Germany 2Max Planck Institute for Chemistry, P.O. Box 3060, 55020 Mainz, Germany 3DOE Joint Genome Institute, Walnut Creek, CA, USA 4University of Colorado, Ecology and Evolutionary Biology, Boulder, CO, USA Correspondence: Viviane R. Després ([email protected]) and Jörn Wehking ([email protected]) Received: 1 December 2017 – Discussion started: 14 December 2017 Revised: 11 June 2018 – Accepted: 15 June 2018 – Published: 11 July 2018 Abstract. Archaea are ubiquitous in terrestrial and marine narchaeaceae, Nitrososphaeraceae, Methanosarcinales, Ther- environments and play an important role in biogeochemical moplasmata, and the genus Nitrosopumilus as the dominating cycles. Although air acts as the primary medium for their taxa. dispersal among different habitats, their diversity and abun- The seasonal dynamics of methanogenic Euryarchaeota dance is not well characterized. The main reason for this point to anthropogenic activities, such as fertilization of agri- lack of insight is that archaea are difficult to culture, seem cultural fields with biogas substrates or manure, as sources of to be low in number in the atmosphere, and have so far been airborne archaea. This study gains a deeper insight into the difficult to detect even with molecular genetic approaches. -

Downloaded from PIR, PRF, Swissprot And
bioRxiv preprint doi: https://doi.org/10.1101/051623; this version posted May 6, 2016. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. Consensus Phylogenetic trees of Fifteen Prokaryotic Aminoacyl-tRNA Synthetase Polypeptides based on Euclidean Geometry of All-Pairs Distances and Concatenation Rhishikesh Bargaje1,2, M. Milner Kumar1,3 and Sohan Prabhakar Modak1* 1Open Vision, Computational Research Laboratory, 759/75 Deccan Gymkhana, Pune 411004, India. 2Institute of Genomics & Integrative Biology, Mall Road, Delhi 110007, India. Computational Research Laboratories Limited, 3rd floor, TACO House, Damle Path, Off Law College Road, Pune-411004, India. Running title: Consensus phylogeny of aminoacyl-tRNA synthetases RB: [email protected] MMK: [email protected] *Corresponding author: SPM: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/051623; this version posted May 6, 2016. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. Abstract Background Most molecular phylogenetic trees depict the relative closeness or the extent of similarity among a set of taxa based on comparison of sequences of homologous genes or proteins. Since the tree topology for individual monogenic traits varies among the same set of organisms and does not overlap taxonomic hierarchy, hence there is a need to generate multidimensional phylogenetic trees.