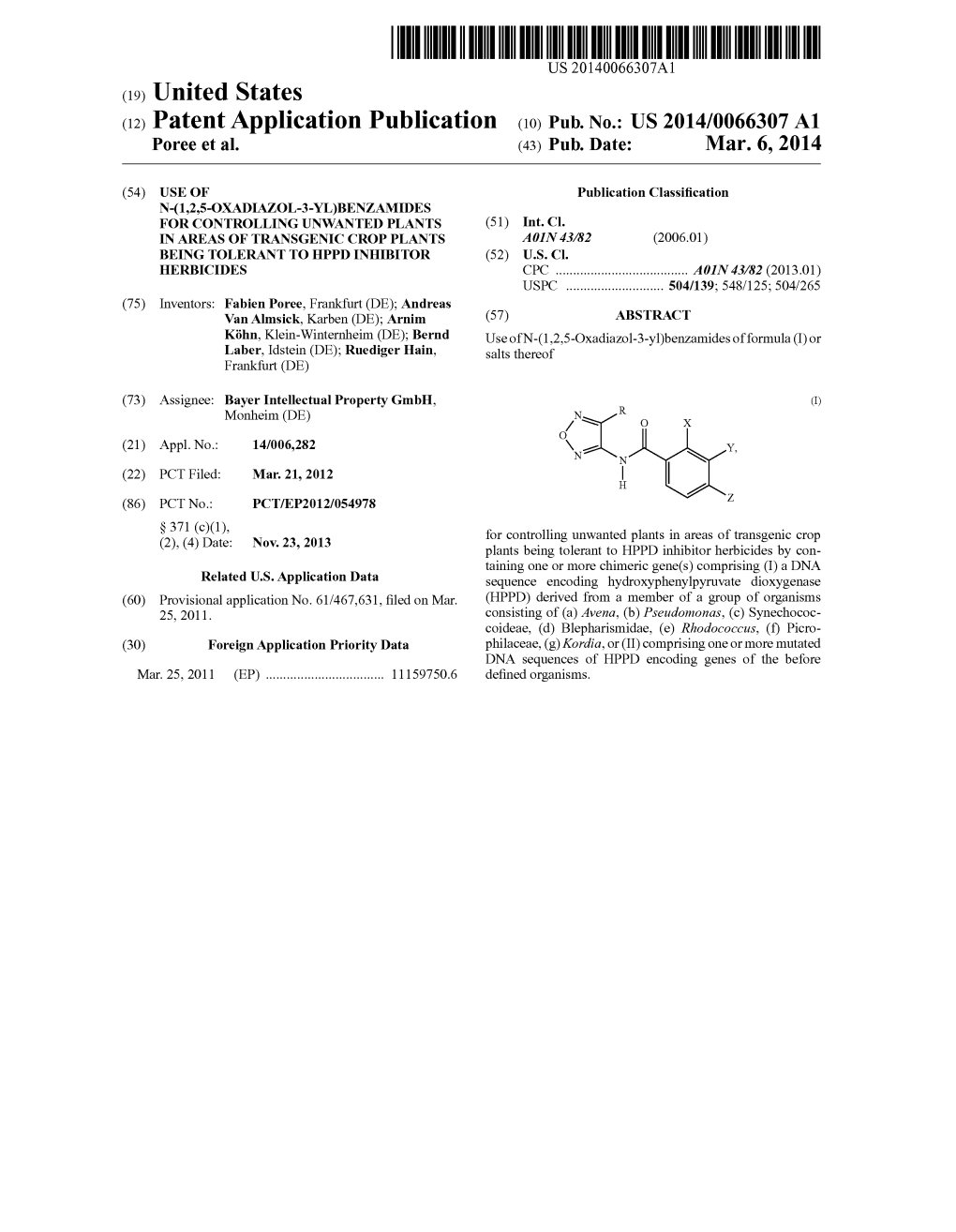
(12) Patent Application Publication (10) Pub. No.: US 2014/0066307 A1 Poree Et Al
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Microbial Diversity of Non-Flooded High Temperature Petroleum Reservoir in South of Iran
Archive of SID Biological Journal of Microorganism th 8 Year, Vol. 8, No. 32, Winter 2020 Received: November 18, 2018/ Accepted: May 21, 2019. Page: 15-231- 8 Microbial Diversity of Non-flooded High Temperature Petroleum Reservoir in South of Iran Mohsen Pournia Department of Microbiology, Shiraz Branch, Islamic Azad University, Shiraz, Iran, [email protected] Nima Bahador * Department of Microbiology, Shiraz Branch, Islamic Azad University, Shiraz, Iran, [email protected] Meisam Tabatabaei Biofuel Research Team (BRTeam), Karaj, Iran, [email protected] Reza Azarbayjani Molecular bank, Iranian Biological Resource Center, ACECR, Karaj, Iran, [email protected] Ghassem Hosseni Salekdeh Department of Biology, Agricultural Biotechnology Research Institute, Karaj, Iran, [email protected] Abstract Introduction: Although bacteria and archaea are able to grow and adapted to the petrol reservoirs during several years, there are no results from microbial diversity of oilfields with high temperature in Iran. Hence, the present study tried to identify microbial community in non-water flooding Zeilaei (ZZ) oil reservoir. Materials and methods: In this study, for the first time, non-water flooded high temperature Zeilaei oilfield was analyzed for its microbial community based on next generation sequencing of 16S rRNA genes. Results: The results obtained from this study indicated that the most abundant bacterial community belonged to phylum of Firmicutes (Bacilli ) and Thermotoga, while other phyla (Proteobacteria , Actinobacteria and Synergistetes ) were much less abundant. Bacillus subtilis , B. licheniformis , Petrotoga mobilis , P. miotherma, Fervidobacterium pennivorans , and Thermotoga subterranea were observed with high frequency. In addition, the most abundant archaea were Methanothermobacter thermautotrophicus . Discussion and conclusion: Although there are many reports on the microbial community of oil filed reservoirs, this is the first report of large quantities of Bacillus spp. -

2I52 Lichtarge Lab 2006
Pages 1–11 2i52 Evolutionary trace report by report maker February 25, 2010 4.3.3 DSSP 10 4.3.4 HSSP 10 4.3.5 LaTex 10 4.3.6 Muscle 10 4.3.7 Pymol 10 4.4 Note about ET Viewer 10 4.5 Citing this work 10 4.6 About report maker 11 4.7 Attachments 11 1 INTRODUCTION From the original Protein Data Bank entry (PDB id 2i52): Title: Crystal structure of protein pto0218 from picrophilus torridus, pfam duf372 Compound: Mol id: 1; molecule: hypothetical protein; chain: a, b, CONTENTS c, d, e, f; engineered: yes Organism, scientific name: Picrophilus Torridus; 1 Introduction 1 2i52 contains a single unique chain 2i52F (119 residues long) and its homologues 2i52A, 2i52D, 2i52C, 2i52E, and 2i52B. 2 Chain 2i52F 1 2.1 Q6L2J9 overview 1 2.2 Multiple sequence alignment for 2i52F 1 2.3 Residue ranking in 2i52F 1 2.4 Top ranking residues in 2i52F and their position on the structure 1 2 CHAIN 2I52F 2.4.1 Clustering of residues at 25% coverage. 1 2.4.2 Overlap with known functional surfaces at 2.1 Q6L2J9 overview 25% coverage. 2 From SwissProt, id Q6L2J9, 97% identical to 2i52F: Description: Hypothetical protein. 3 Notes on using trace results 9 Organism, scientific name: Picrophilus torridus. 3.1 Coverage 9 Taxonomy: Archaea; Euryarchaeota; Thermoplasmata; Thermoplas- 3.2 Known substitutions 9 matales; Picrophilaceae; Picrophilus. 3.3 Surface 9 3.4 Number of contacts 9 3.5 Annotation 9 3.6 Mutation suggestions 9 2.2 Multiple sequence alignment for 2i52F 4 Appendix 9 For the chain 2i52F, the alignment 2i52F.msf (attached) with 30 4.1 File formats 9 sequences was used. -

Proteome Cold-Shock Response in the Extremely Acidophilic Archaeon, Cuniculiplasma Divulgatum
microorganisms Article Proteome Cold-Shock Response in the Extremely Acidophilic Archaeon, Cuniculiplasma divulgatum Rafael Bargiela 1 , Karin Lanthaler 1,2, Colin M. Potter 1,2 , Manuel Ferrer 3 , Alexander F. Yakunin 1,2, Bela Paizs 1,2, Peter N. Golyshin 1,2 and Olga V. Golyshina 1,2,* 1 School of Natural Sciences, Bangor University, Deiniol Rd, Bangor LL57 2UW, UK; [email protected] (R.B.); [email protected] (K.L.); [email protected] (C.M.P.); [email protected] (A.F.Y.); [email protected] (B.P.); [email protected] (P.N.G.) 2 Centre for Environmental Biotechnology, Bangor University, Deiniol Rd, Bangor LL57 2UW, UK 3 Systems Biotechnology Group, Department of Applied Biocatalysis, CSIC—Institute of Catalysis, Marie Curie 2, 28049 Madrid, Spain; [email protected] * Correspondence: [email protected]; Tel.: +44-1248-388607; Fax: +44-1248-382569 Received: 27 April 2020; Accepted: 15 May 2020; Published: 19 May 2020 Abstract: The archaeon Cuniculiplasma divulgatum is ubiquitous in acidic environments with low-to-moderate temperatures. However, molecular mechanisms underlying its ability to thrive at lower temperatures remain unexplored. Using mass spectrometry (MS)-based proteomics, we analysed the effect of short-term (3 h) exposure to cold. The C. divulgatum genome encodes 2016 protein-coding genes, from which 819 proteins were identified in the cells grown under optimal conditions. In line with the peptidolytic lifestyle of C. divulgatum, its intracellular proteome revealed the abundance of proteases, ABC transporters and cytochrome C oxidase. From 747 quantifiable polypeptides, the levels of 582 proteins showed no change after the cold shock, whereas 104 proteins were upregulated suggesting that they might be contributing to cold adaptation. -

Insights Into Archaeal Evolution and Symbiosis from the Genomes of a Nanoarchaeon and Its Inferred Crenarchaeal Host from Obsidian Pool, Yellowstone National Park
University of Tennessee, Knoxville TRACE: Tennessee Research and Creative Exchange Microbiology Publications and Other Works Microbiology 4-22-2013 Insights into archaeal evolution and symbiosis from the genomes of a nanoarchaeon and its inferred crenarchaeal host from Obsidian Pool, Yellowstone National Park Mircea Podar University of Tennessee - Knoxville, [email protected] Kira S. Makarova National Institutes of Health David E. Graham University of Tennessee - Knoxville, [email protected] Yuri I. Wolf National Institutes of Health Eugene V. Koonin National Institutes of Health See next page for additional authors Follow this and additional works at: https://trace.tennessee.edu/utk_micrpubs Part of the Microbiology Commons Recommended Citation Biology Direct 2013, 8:9 doi:10.1186/1745-6150-8-9 This Article is brought to you for free and open access by the Microbiology at TRACE: Tennessee Research and Creative Exchange. It has been accepted for inclusion in Microbiology Publications and Other Works by an authorized administrator of TRACE: Tennessee Research and Creative Exchange. For more information, please contact [email protected]. Authors Mircea Podar, Kira S. Makarova, David E. Graham, Yuri I. Wolf, Eugene V. Koonin, and Anna-Louise Reysenbach This article is available at TRACE: Tennessee Research and Creative Exchange: https://trace.tennessee.edu/ utk_micrpubs/44 Podar et al. Biology Direct 2013, 8:9 http://www.biology-direct.com/content/8/1/9 RESEARCH Open Access Insights into archaeal evolution and symbiosis from the genomes of a nanoarchaeon and its inferred crenarchaeal host from Obsidian Pool, Yellowstone National Park Mircea Podar1,2*, Kira S Makarova3, David E Graham1,2, Yuri I Wolf3, Eugene V Koonin3 and Anna-Louise Reysenbach4 Abstract Background: A single cultured marine organism, Nanoarchaeum equitans, represents the Nanoarchaeota branch of symbiotic Archaea, with a highly reduced genome and unusual features such as multiple split genes. -

Picrophilus Oshimae and Picrophilus Tomdus Fam. Nov., Gen. Nov., Sp. Nov
INTERNATIONALJOURNAL OF SYSTEMATICBACTERIOLOGY, July 1996, p. 814-816 Vol. 46, No. 3 0020-77 13/96/$04.00+0 Copyright 0 1996, International Union of Microbiological Societies Picrophilus oshimae and Picrophilus tomdus fam. nov., gen. nov., sp. nov., Two Species of Hyperacidophilic, Thermophilic, Heterotrophic, Aerobic Archaea CHRISTA SCHLEPER, GABRIELA PUHLER, HANS- PETER KLENK, AND WOLFRAM ZILLIG* Max Plank Institut fur Biochemie, 0-82152 Martinsried, Germany We describe two species of hyperacidophilic, thermophilic, heterotrophic, aerobic archaea that were isolated from solfataric hydrothermal areas in Hokkaido, Japan. These organisms, Picrophilus oshimae and Picrophilus torridus, represent a novel genus and a novel family, the Picrophilaceae, in the kingdom Euryarchaeota and the order Thermoplasmales. Both of these bacteria are more acidophilic than the genus Thermoplasma since they are able to grow at about pH 0. The moderately thermophilic, hyperacidophilic, aerobic ar- which comprises acid-loving (i.e., hyperacidophilic) organisms. chaea (archaebacteria) (7) Picrophilus oshimae and Picrophilus Separation of these taxa is justified by their phylogenetic dis- rorridus, which have been described previously (4, 5), were tance, (9.5% difference in the 16s rRNA sequences of mem- isolated from moderately hot hydrothermal areas in solfataric bers of the Picrophilaceae and T. acidophilum), by the lack of fields in Hokkaido, Japan. One of the sources of isolation was immunochemical cross-reactions in Ouchterlony immunodif- a solfataric spring which had a temperature of 53°C and a pH fusion assays between the RNA polymerases of P. oshimae and of 2.2, and the other was a rather dry hot soil which had a pH T. acidophilum, which also do not occur between members of of <OS. -

Downloaded from PIR, PRF, Swissprot And
bioRxiv preprint doi: https://doi.org/10.1101/051623; this version posted May 6, 2016. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. Consensus Phylogenetic trees of Fifteen Prokaryotic Aminoacyl-tRNA Synthetase Polypeptides based on Euclidean Geometry of All-Pairs Distances and Concatenation Rhishikesh Bargaje1,2, M. Milner Kumar1,3 and Sohan Prabhakar Modak1* 1Open Vision, Computational Research Laboratory, 759/75 Deccan Gymkhana, Pune 411004, India. 2Institute of Genomics & Integrative Biology, Mall Road, Delhi 110007, India. Computational Research Laboratories Limited, 3rd floor, TACO House, Damle Path, Off Law College Road, Pune-411004, India. Running title: Consensus phylogeny of aminoacyl-tRNA synthetases RB: [email protected] MMK: [email protected] *Corresponding author: SPM: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/051623; this version posted May 6, 2016. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. Abstract Background Most molecular phylogenetic trees depict the relative closeness or the extent of similarity among a set of taxa based on comparison of sequences of homologous genes or proteins. Since the tree topology for individual monogenic traits varies among the same set of organisms and does not overlap taxonomic hierarchy, hence there is a need to generate multidimensional phylogenetic trees. -

Occurrence and Expression of Novel Methyl-Coenzyme M Reductase Gene
www.nature.com/scientificreports OPEN Occurrence and expression of novel methyl-coenzyme M reductase gene (mcrA) variants in hot spring Received: 6 April 2017 Accepted: 27 June 2017 sediments Published: xx xx xxxx Luke J. McKay1,2, Roland Hatzenpichler1,3, William P. Inskeep2 & Matthew W. Fields1,4 Recent discoveries have shown that the marker gene for anaerobic methane cycling (mcrA) is more widespread in the Archaea than previously thought. However, it remains unclear whether novel mcrA genes associated with the Bathyarchaeota and Verstraetearchaeota are distributed across diverse environments. We examined two geochemically divergent but putatively methanogenic regions of Yellowstone National Park to investigate whether deeply-rooted archaea possess and express novel mcrA genes in situ. Small-subunit (SSU) rRNA gene analyses indicated that Bathyarchaeota were predominant in seven of ten sediment layers, while the Verstraetearchaeota and Euryarchaeota occurred in lower relative abundance. Targeted amplifcation of novel mcrA genes suggested that diverse taxa contribute to alkane cycling in geothermal environments. Two deeply-branching mcrA clades related to Bathyarchaeota were identifed, while highly abundant verstraetearchaeotal mcrA sequences were also recovered. In addition, detection of SSU rRNA and mcrA transcripts from one hot spring suggested that predominant Bathyarchaeota were also active, and that methane cycling genes are expressed by the Euryarchaeota, Verstraetearchaeota, and an unknown lineage basal to the Bathyarchaeota. These fndings greatly expand the diversity of the key marker gene for anaerobic alkane cycling and outline the need for greater understanding of the functional capacity and phylogenetic afliation of novel mcrA variants. Archaea are the primary drivers of CH4 cycling on our planet. -

(Gid ) Genes Coding for Putative Trna:M5u-54 Methyltransferases in 355 Bacterial and Archaeal Complete Genomes
Table S1. Taxonomic distribution of the trmA and trmFO (gid ) genes coding for putative tRNA:m5U-54 methyltransferases in 355 bacterial and archaeal complete genomes. Asterisks indicate the presence and the number of putative genes found. Genomes Taxonomic position TrmA Gid Archaea Crenarchaea Aeropyrum pernix_K1 Crenarchaeota; Thermoprotei; Desulfurococcales; Desulfurococcaceae Cenarchaeum symbiosum Crenarchaeota; Thermoprotei; Cenarchaeales; Cenarchaeaceae Pyrobaculum aerophilum_str_IM2 Crenarchaeota; Thermoprotei; Thermoproteales; Thermoproteaceae Sulfolobus acidocaldarius_DSM_639 Crenarchaeota; Thermoprotei; Sulfolobales; Sulfolobaceae Sulfolobus solfataricus Crenarchaeota; Thermoprotei; Sulfolobales; Sulfolobaceae Sulfolobus tokodaii Crenarchaeota; Thermoprotei; Sulfolobales; Sulfolobaceae Euryarchaea Archaeoglobus fulgidus Euryarchaeota; Archaeoglobi; Archaeoglobales; Archaeoglobaceae Haloarcula marismortui_ATCC_43049 Euryarchaeota; Halobacteria; Halobacteriales; Halobacteriaceae; Haloarcula Halobacterium sp Euryarchaeota; Halobacteria; Halobacteriales; Halobacteriaceae; Haloarcula Haloquadratum walsbyi Euryarchaeota; Halobacteria; Halobacteriales; Halobacteriaceae; Haloquadra Methanobacterium thermoautotrophicum Euryarchaeota; Methanobacteria; Methanobacteriales; Methanobacteriaceae Methanococcoides burtonii_DSM_6242 Euryarchaeota; Methanomicrobia; Methanosarcinales; Methanosarcinaceae Methanococcus jannaschii Euryarchaeota; Methanococci; Methanococcales; Methanococcaceae Methanococcus maripaludis_S2 Euryarchaeota; Methanococci; -

The Novel Extremely Acidophilic, Cell-Wall-Deficient Archaeon Cuniculiplasma Divulgatum Gen
International Journal of Systematic and Evolutionary Microbiology (2016), 66, 332–340 DOI 10.1099/ijsem.0.000725 The novel extremely acidophilic, cell-wall-deficient archaeon Cuniculiplasma divulgatum gen. nov., sp. nov. represents a new family, Cuniculiplasmataceae fam. nov., of the order Thermoplasmatales Olga V. Golyshina,1 Heinrich Lu¨nsdorf,2 Ilya V. Kublanov,3 Nadine I. Goldenstein,4 Kai-Uwe Hinrichs4 and Peter N. Golyshin1 Correspondence 1School of Biological Sciences, Bangor University, Deiniol Rd, Bangor LL57 2UW, UK Olga V. Golyshina 2Central Unit of Microscopy, Helmholtz Centre for Infection Research, Inhoffenstrasse 7, [email protected] Braunschweig 38124, Germany 3Winogradsky Institute of Microbiology, Russian Academy of Sciences, Prospect 60-Letiya Oktyabrya 7/2, Moscow 117312, Russia 4MARUM – Center for Marine Environmental Sciences, University of Bremen, Leobener Str., Bremen 28359, Germany Two novel cell-wall-less, acidophilic, mesophilic, organotrophic and facultatively anaerobic archaeal strains were isolated from acidic streamers formed on the surfaces of copper-ore- containing sulfidic deposits in south-west Spain and North Wales, UK. Cells of the strains varied from 0.1 to 2 mm in size and were pleomorphic, with a tendency to form filamentous structures. The optimal pH and temperature for growth for both strains were 1.0–1.2 and 2 37–40 8C, with the optimal substrates for growth being beef extract (3 g l 1) for strain S5T and 2 beef extract with tryptone (3 and 1 g l 1, respectively) for strain PM4. The lipid composition was dominated by intact polar lipids consisting of a glycerol dibiphytanyl glycerol tetraether (GDGT) core attached to predominantly glycosidic polar headgroups. -

Microbial Extremophiles in Aspect of Limits of Life. Elena V. ~Ikuta
Source of Acquisition NASA Marshall Space Flight Centel Microbial extremophiles in aspect of limits of life. Elena V. ~ikuta',Richard B. ~oover*,and Jane an^.^ IT LF " '-~ationalSpace Sciences and Technology CenterINASA, VP-62, 320 Sparkman Dr., Astrobiology Laboratory, Huntsville, AL 35805, USA. 3- Noblis, 3 150 Fairview Park Drive South, Falls Church, VA 22042, USA. Introduction. During Earth's evolution accompanied by geophysical and climatic changes a number of ecosystems have been formed. These ecosystems differ by the broad variety of physicochemical and biological factors composing our environment. Traditionally, pH and salinity are considered as geochemical extremes, as opposed to the temperature, pressure and radiation that are referred to as physical extremes (Van den Burg, 2003). Life inhabits all possible places on Earth interacting with the environment and within itself (cross species relations). In nature it is very rare when an ecotope is inhabited by a single species. As a rule, most ecosystems contain the functionally related and evolutionarily adjusted communities (consortia and populations). In contrast to the multicellular structure of eukaryotes (tissues, organs, systems of organs, whole organism), the highest organized form of prokaryotic life in nature is the benthic colonization in biofilms and microbial mats. In these complex structures all microbial cells of different species are distributed in space and time according to their functions and to physicochemical gradients that allow more effective system support, self- protection, and energy distribution. In vitro, of course, the most primitive organized structure for bacterial and archaeal cultures is the colony, the size, shape, color, consistency, and other characteristics of which could carry varies specifics on species or subspecies levels. -

Extremophiles-Basic Concepts
CONTENTS CONTENTS EXTREMOPHILES Extremophiles - Volume 1 No. of Pages: 396 ISBN: 978-1-905839-93-3 (eBook) ISBN: 978-1-84826-993-4 (Print Volume) Extremophiles - Volume 2 No. of Pages: 392 ISBN: 978-1-905839-94-0 (eBook) ISBN: 978-1-84826-994-1 (Print Volume) Extremophiles - Volume 3 No. of Pages: 364 ISBN: 978-1-905839-95-7 (eBook) ISBN: 978-1-84826-995-8 (Print Volume) For more information of e-book and Print Volume(s) order, please click here Or contact : [email protected] ©Encyclopedia of Life Support Systems (EOLSS) EXTREMOPHILES CONTENTS VOLUME I Extremophiles: Basic Concepts 1 Charles Gerday, Laboratory of Biochemistry, University of Liège, Belgium 1. Introduction 2. Effects of Extreme Conditions on Cellular Components 2.1. Membrane Structure 2.2. Nucleic Acids 2.2.1. Introduction 2.2.2. Desoxyribonucleic Acids 2.2.3. Ribonucleic Acids 2.3. Proteins 2.3.1. Introduction 2.3.2. Thermophilic Proteins 2.3.2.1. Enthalpically Driven Stabilization Factors: 2.3.2.2. Entropically Driven Stabilization Factors: 2.3.3. Psychrophilic Proteins 2.3.4. Halophilic Proteins 2.3.5. Piezophilic Proteins 2.3.5.1. Interaction with Other Proteins and Ligands: 2.3.5.2. Substrate Binding and Catalytic Efficiency: 2.3.6. Alkaliphilic Proteins 2.3.7. Acidophilic Proteins 3. Conclusions Extremophiles: Overview of the Biotopes 43 Michael Gross, University of London, London, UK 1. Introduction 2. Extreme Temperatures 2.1. Terrestrial Hot Springs 2.2. Hot Springs on the Ocean Floor and Black Smokers 2.3. Life at Low Temperatures 3. High Pressure 3.1. -

Distribution and Evolution of the Mobile Vma-1B Intein
Authors' pre-print version. Manuscript published in Molecular Biology and Evolution MBE-13-0254.R2 DOI of published manuscript: 10.1093/molbev/mst164 Article Discoveries Section Distribution and Evolution of the Mobile vma-1b Intein. Kristen S. Swithers1,3, Shannon M. Soucy1, Erica Lasek-Nesselquist1,5, Pascal Lapierre2,4 and Johann Peter Gogarten1* 1 Department of Molecular and Cell Biology, University of Connecticut, Storrs, Connecticut, United States of America 2 University of Connecticut Biotechnology Center, University of Connecticut, Storrs, Connecticut, United States of America 3 Current affiliation: Department of Cell Biology, Yale University Medical School, New Haven, Connecticut, United States of America 4 Current affiliation: New York State Department of Health, Wadsworth Center, Albany, New York, United States of America 5 Current affiliation: Department of Biology, University of Scranton, Scranton, PA, United States of America Communicating author: Johann Peter Gogarten Communicating author email: [email protected], [email protected] !"#"!" Abstract Inteins are self-splicing parasitic genetic elements found in all domains of life. These genetic elements are found in highly conserved positions in conserved proteins. One protein family that has been invaded by inteins is the vacuolar and archaeal catalytic ATPase subunits (vma-1). There are two intein insertion sites in this protein, "a" and "b". The "b" site was previously thought to be only invaded in archaeal lineages. Here we survey the distribution and evolutionary histories of the "b" site inteins and show the intein is present in more lineages than previously annotated including a bacterial lineage, Mahalla australienses 50-1 BON. We present evidence, through ancestral character state reconstruction and substitution ratios between host genes and inteins, for several transfers of this intein between divergent species, including an interdomain transfer between the Archaea and Bacteria.