Accurate Estimation of Cell-Type Composition from Gene Expression Data
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Introduction to the Cell Cell History Cell Structures and Functions
Introduction to the cell cell history cell structures and functions CK-12 Foundation December 16, 2009 CK-12 Foundation is a non-profit organization with a mission to reduce the cost of textbook materials for the K-12 market both in the U.S. and worldwide. Using an open-content, web-based collaborative model termed the “FlexBook,” CK-12 intends to pioneer the generation and distribution of high quality educational content that will serve both as core text as well as provide an adaptive environment for learning. Copyright ©2009 CK-12 Foundation This work is licensed under the Creative Commons Attribution-Share Alike 3.0 United States License. To view a copy of this license, visit http://creativecommons.org/licenses/by-sa/3.0/us/ or send a letter to Creative Commons, 171 Second Street, Suite 300, San Francisco, California, 94105, USA. Contents 1 Cell structure and function dec 16 5 1.1 Lesson 3.1: Introduction to Cells .................................. 5 3 www.ck12.org www.ck12.org 4 Chapter 1 Cell structure and function dec 16 1.1 Lesson 3.1: Introduction to Cells Lesson Objectives • Identify the scientists that first observed cells. • Outline the importance of microscopes in the discovery of cells. • Summarize what the cell theory proposes. • Identify the limitations on cell size. • Identify the four parts common to all cells. • Compare prokaryotic and eukaryotic cells. Introduction Knowing the make up of cells and how cells work is necessary to all of the biological sciences. Learning about the similarities and differences between cell types is particularly important to the fields of cell biology and molecular biology. -
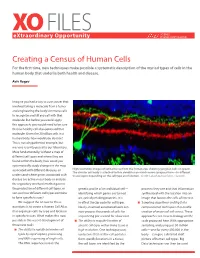
Creating a Census of Human Cells
XO FILES SCIENCE eXtraordinary Opportunity PHILANTHROPY ALLIANCE Creating a Census of Human Cells For the first time, new techniques make possible a systematic description of the myriad types of cells in the human body that underlie both health and disease. Aviv Regev Imagine you had a way to cure cancer that involved taking a molecule from a tumor and engineering the body’s immune cells to recognize and kill any cell with that molecule. But before you could apply this approach, you would need to be sure that no healthy cell also expressed that molecule. Given the 20 trillion cells in a human body, how would you do that? This is not a hypothetical example, but was one recently posed to our laboratory. More fundamentally, without a map of different cell types and where they are found within the body, how could you systematically study changes in the map associated with different diseases, or High resolution images of retinal tissue from the human eye, showing ganglion cells (in green). The circular cell body is attached to thin dendrites on which nerve synapses form—in different understand where genes associated with tissue layers depending on the cell type and function. Credit: Lab of Joshua Sanes, Harvard. disease are active in our body or analyze the regulatory mechanisms that govern the production of different cell types, or genetic profile of an individual cell— proteins they use and that information sort out how different cell types combine identifying which genes are turned synthesized with the location into an to form specific tissues? on, actively making proteins. -

Robustness and Timing of Cellular Differentiation Through Population-Based Symmetry Breaking
AR TI CLE Robustness AND TIMING OF CELLULAR DIFFERENTIATION THROUGH population-based SYMMETRY BREAKING Angel StanoeV1 | Christian Schröter1 | Aneta KOSESKA1∗ 1Department OF Systemic Cell Biology, Max Planck Institute OF Molecular PhYSIOLOGY, Although COORDINATED BEHAVIOR OF MULTICELLULAR ORGANISMS Dortmund, GermanY RELIES ON INTERCELLULAR communication, THE DYNAMICAL mech- Correspondence ANISM THAT UNDERLIES SYMMETRY BREAKING IN HOMOGENEOUS Email: populations, THEREBY GIVING RISE TO HETEROGENEOUS CELL TYPES ∗[email protected] REMAINS UNCLEAR. In CONTRAST TO THE PREVALENT cell-intrinsic VIEW OF DIFFERENTIATION WHERE MULTISTABILITY ON THE SINGLE CELL LEVEL IS A NECESSARY pre-requisite, WE PROPOSE THAT het- EROGENEOUS CELLULAR IDENTITIES EMERGE AND ARE MAINTAINED COOPERATIVELY ON A POPULATION LEVel. Identifying A GENERIC SYMMETRY BREAKING mechanism, WE DEMONSTRATE THAT ROBUST PROPORTIONS BETWEEN DIFFERENTIATED CELL TYPES ARE INTRINSIC FEATURES OF THIS INHOMOGENEOUS STEADY STATE solution. Crit- ICAL ORGANIZATION IN THE VICINITY OF THE SYMMETRY BREAKING BIFURCATION ON THE OTHER hand, SUGGESTS THAT TIMING OF cel- LULAR DIFFERENTIATION EMERGES FOR GROWING POPULATIONS IN A self-organized MANNER. Setting A CLEAR DISTINCTION BETWEEN pre-eXISTING ASYMMETRIES AND SYMMETRY BREAKING EVents, WE THUS DEMONSTRATE THAT EMERGENT SYMMETRY breaking, IN CONJUNCTION WITH ORGANIZATION NEAR CRITICALITY NECESSARILY LEADS TO ROBUSTNESS AND ACCURACY DURING DEVelopment. KEYWORDS SYMMETRY breaking; differentiation; INHOMOGENEOUS STEADY STATE Abbreviations: -

A Concise Review on the Classification and Nomenclature of Stem Cells Kök Hücrelerinin S›N›Fland›R›Lmas› Ve Isimlendirilmesine Iliflkin K›Sa Bir Derleme
Review 57 A concise review on the classification and nomenclature of stem cells Kök hücrelerinin s›n›fland›r›lmas› ve isimlendirilmesine iliflkin k›sa bir derleme Alp Can Ankara University Medical School, Department of Histology and Embryology, Ankara, Turkey Abstract Stem cell biology and regenerative medicine is a relatively young field. However, in recent years there has been a tremen- dous interest in stem cells possibly due to their therapeutic potential in disease states. As a classical definition, a stem cell is an undifferentiated cell that can produce daughter cells that can either remain a stem cell in a process called self-renew- al, or commit to a specific cell type via the initiation of a differentiation pathway leading to the production of mature progeny cells. Despite this acknowledged definition, the classification of stem cells has been a perplexing notion that may often raise misconception even among stem cell biologists. Therefore, the aim of this brief review is to give a conceptual approach to classifying the stem cells beginning from the early morula stage totipotent embryonic stem cells to the unipotent tissue-resident adult stem cells, also called tissue-specific stem cells. (Turk J Hematol 2008; 25: 57-9) Key words: Stem cells, embryonic stem cells, tissue-specific stem cells, classification, progeny. Özet Kök hücresi biyolojisi ve onar›msal t›p görece yeni alanlard›r. Buna karfl›n, son y›llarda çeflitli hastal›klarda tedavi amac›yla kullan›labilme potansiyelleri nedeniyle kök hücrelerine ola¤anüstü bir ilgi art›fl› vard›r. Klasik tan›m›yla kök hücresi, kendini yenileme ad› verilen mekanizmayla farkl›laflmadan kendini ço¤altan veya bir dizi farkl›laflma aflamas›ndan geçerek olgun hücrelere dönüflebilen hücrelerdir. -

An Introduction to Stem Cell Biology
An Introduction to Stem Cell Biology Michael L. Shelanski, MD,PhD Professor of Pathology and Cell Biology Columbia University Figures adapted from ISSCR. Presentations of Drs. Martin Pera (Monash University), Dr.Susan Kadereit, Children’s Hospital, Boston and Dr. Catherine Verfaillie, University of Minnesota Science 1999, 283: 534-537 PNAS 1999, 96: 14482-14486 Turning Blood into Brain: Cells Bearing Neuronal Antigens Generated in Vitro from Bone Marrow Science 2000, 290:1779-1782 From Marrow to Brain: Expression of Neuronal Phenotypes in Adult Mice Mezey, E., Chandross, K.J., Harta, G., Maki, R.A., McKercher, S.R. Science 2000, 290:1775-1779 Brazelton, T.R., Rossi, F.M., Keshet, G.I., Blau, H.M. Nature 2001, 410:701-705 Nat Med 2000, 11: 1229-1234 Stem Cell FAQs Do you need to get one from an egg? Must you sacrifice an Embryo? What is an ES cell? What about adult stem cells or cord blood stem cells Why can’t this work be done in animals? Are “cures” on the horizon? Will this lead to human cloning – human spare parts factories? Are we going to make a Frankenstein? What is a stem cell? A primitive cell which can either self renew (reproduce itself) or give rise to more specialised cell types The stem cell is the ancestor at the top of the family tree of related cell types. One blood stem cell gives rise to red cells, white cells and platelets Stem Cells Vary in their Developmental capacity A multipotent cell can give rise to several types of mature cell A pluripotent cell can give rise to all types of adult tissue cells plus extraembryonic tissue: cells which support embryonic development A totipotent cell can give rise to a new individual given appropriate maternal support The Fertilized Egg The “Ultimate” Stem Cell – the Newly Fertilized Egg (one Cell) will give rise to all the cells and tissues of the adult animal. -

Stem and Cancer Stem Cell Identities, Cellular Markers, Niche Environment and Response to Treatments to Unravel New Therapeutic Targets
biology Editorial Stem and Cancer Stem Cell Identities, Cellular Markers, Niche Environment and Response to Treatments to Unravel New Therapeutic Targets Jose R. Pineda 1,2,* , Iker Badiola 1 and Gaskon Ibarretxe 1,* 1 Department of Cell Biology and Histology, Faculty of Medicine and Nursing, University of the Basque Country (UPV/EHU), 48940 Leioa, Spain; [email protected] 2 Achucarro Basque Center for Neuroscience Fundazioa, 48940 Leioa, Spain * Correspondence: [email protected] (J.R.P.); [email protected] (G.I.); Tel.: +34-9460-12426 (J.R.P.); +34-9460-13218 (G.I.) Adult stem cells are a partially quiescent cell population responsible for natural cell renewal and are found in many different regions of the body, including the brain, teeth, bones, muscles, skin, and diverse epithelia, such as the epidermal or intestinal epithelium, among others. Interestingly, adult stem cell populations share all the instructions to grow and differentiate to any type of cell of the specific tissue they belong to. These normally quiescent stem cells can be activated on demand to replenish mature differentiated cell populations, a process which is driven mainly by signaling cues from its niche, thanks to the activation of their specific receptors in a timely and fine-tuned manner. One of the best examples of a very different niche regulation is the radically different turnover rate of intestinal stem cells with respect to neural stem cells. When someone evokes the word “cancer”, it can inspire fear and respect for a devas- Citation: Pineda, J.R.; Badiola, I.; tating and rampant disease, and similarly it could give the impression at first glance that it Ibarretxe, G. -

LECTURE NOTES : CELL BIOLOGY Author(S): Dr
LECTURE NOTES: CELL BIOLOGY (BIOMEDICAL LABORATORY SCIENCE STUDENTS) By Dr. Callixte Yadufashije Senior lecturer INES Ruhengeri ( Ruhengeri Institute Of Higher Education) January, 2018 2018 Ideal International E – Publication Pvt. Ltd. www.isca.co.in 427, Palhar Nagar, RAPTC, VIP-Road, Indore-452005 (MP) INDIA Phone: +91-731-2616100, Mobile: +91-80570-83382 E-mail: [email protected] , Website: www.isca.co.in Title: LECTURE NOTES : CELL BIOLOGY Author(s): Dr. Callixte Yadufashije Edition: First Volume: I © Copyright Reserved 2018 All rights reserved. No part of this publication may be reproduced, stored, in a retrieval system or transmitted, in any form or by any means, electronic, mechanical, photocopying, reordering or otherwise, without the prior permission of the publisher. ISBN: 978-93-86675-40-8 Ideal International E- Publication www.isca.co.in iii Preface Cell biology also known as cytology. It deals with cells and its organelles. Inside this document you find curious of yours about cells and how they act like independent livings within bodies of living organisms. Preparation of this lecture notes has took much effort as needs high understanding in science of biology. Cells are important parts of life and without them life can be impossible. Understanding sciences like cytology, histology, genetics, biochemistry, molecular biology, are essential sciences to know are linked to knowledge of cells. During preparation of this lecture notes, content of module of cell biology taught in biomedical sciences courses in INES-Ruhengeri at undergraduate level has been used. A part from this content different source has been used to study each topic with much attention. -

FUT9-Driven Programming of Colon Cancer Cells Towards a Stem Cell-Like State
cancers Article FUT9-Driven Programming of Colon Cancer Cells towards a Stem Cell-Like State Athanasios Blanas y, Anouk Zaal y, Irene van der Haar Àvila , Maxime Kempers , Laura Kruijssen, Mike de Kok , Marko A. Popovic, Joost C. van der Horst and Sandra J. van Vliet * Department of Molecular Cell Biology and Immunology, Amsterdam Infection & Immunity Institute, Cancer Center Amsterdam, Vrije Universiteit Amsterdam, Amsterdam UMC, 1081 HZ Amsterdam, The Netherlands; [email protected] (A.B.); [email protected] (A.Z.); [email protected] (I.v.d.H.À.); [email protected] (M.K.); [email protected] (L.K.); [email protected] (M.d.K.); [email protected] (M.A.P.); [email protected] (J.C.v.d.H.) * Correspondence: [email protected]; Tel.: +31-20-4448080 Equal contribution. y Received: 9 June 2020; Accepted: 1 September 2020; Published: 10 September 2020 Simple Summary: Aberrant glycosylation, for instance heightened expression of fucosylated structures, is a frequent feature observed in tumor cells. Our paper outlines the role of aberrant fucosylation by the Fucosyltransferase 9 (FUT9) as a potent reprogramming factor marking the acquisition of a stem-like state both by murine and human colon cancer cells. Importantly, our study reinforces the implication of aberrant fucosylation in promoting tumor growth and resistance to chemotherapy in the context of colon cancer. Abstract: Cancer stem cells (CSCs) are located in dedicated niches, where they remain inert to chemotherapeutic drugs and drive metastasis. Although plasticity in the CSC pool is well appreciated, the molecular mechanisms implicated in the regulation of cancer stemness are still elusive. -

Studing Cells
Studing Cells What are cells? Cells provide structure and function for all living things, from microorganisms to humans. Scientists consider them the smallest form of life. Cells house the biological machinery that makes the proteins, chemicals, and signals responsible for everything that happens inside our bodies. What do cells look like? Cells come in different shapes—round, flat, long, star-like, cubed, and even shapeless. Most cells are colorless and see-through. The size of a cell also varies. Some of the smallest are one-celled bacteria, which are too small to see with the naked eye, at 1-millionth of a meter (micrometer) across. Plants have some of the largest cells, 10–100 micrometers across. The human cell with the biggest diameter is the egg. It is about the same diameter as a hair strand (80 micrometers). How many different types of human cells are there? Fibroblast cells with nuclei (blue, circular, center), energy factories The trillions of cells that make up a human are organized into about 200 major (green, surrounding the nucleus), and types. All of a person’s cells contain the same set of genes (learn more on the the actin cytoskeleton (red, outermost). Credit: Dylan Burnette and Jennifer Genetics Home Reference What is a gene? webpage). However, each cell type Lippincott-Schwartz, Eunice Kennedy “switches on” a different pattern of genes, and this determines which proteins the Shriver National Institute of Child Health cell produces. The unique set of proteins in different cell types allows them to and Human Development, National perform specialized tasks. For instance, red blood cells carry oxygen throughout the Institutes of Health. -

Identification of Cell-Type-Specific Marker Genes from Co-Expression Patterns in Tissue Samples
bioRxiv preprint doi: https://doi.org/10.1101/2020.11.07.373043; this version posted November 8, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Identification of cell-type-specific marker genes from co-expression patterns in tissue samples Yixuan Qiu 1, Jiebiao Wang 2, Jing Lei 1, and Kathryn Roeder 1;3 1 Department of Statistics and Data Science, Carnegie Mellon University, USA 2 Department of Biostatistics, University of Pittsburgh, USA 3 Computational Biology Department, Carnegie Mellon University, USA Abstract Motivation: Marker genes, defined as genes that are expressed primarily in a single cell type, can be identified from the single cell transcriptome; however, such data are not always available for the many uses of marker genes, such as deconvolution of bulk tissue. Marker genes for a cell type, however, are highly correlated in bulk data, because their expression levels depend primarily on the proportion of that cell type in the samples. Therefore, when many tissue samples are analyzed, it is possible to identify these marker genes from the correlation pattern. Results: To capitalize on this pattern, we develop a new algorithm to detect marker genes by combining published information about likely marker genes with bulk transcriptome data in the form of a semi-supervised algorithm. The al- gorithm then exploits the correlation structure of the bulk data to refine the published marker genes by adding or removing genes from the list. -

17 Cell Differentiation and Gene Expression Investigation a N D M O D E L I N G • 1–2 C L a S S S E S S I O N S
17 Cell Differentiation and Gene Expression investigation a n d m o d e l i n g • 1–2 c l a s s s e s s i o n s ACTIVITY OVERVIEW TeachIng summary Students investigate gene expression as it relates to cell Getting Started differentiation in four human cell types. They consider how • Elicit students’ ideas about the genetic makeup of various physiological events affect gene expression in each different cells in a multicellular organism. of the four cell types. Doing the Activity Key COntenT • Students investigate gene expression. 1. The expression of specific genes regulates cell differen Follow-up tiation and cell functions. • The class discusses gene expression and gene regulation. 2. Somatic cells in an individual organism have the same • (LITERACY) Students read a case study about terminator genome, but selectively express the genes for production genes. of characteristic proteins. 3. The proteins a cell produces determine that cell’s Background Information phenotype. Gene Expression Gene expression is the process in which the information KEY Process sKIlls stored in DNA is used to produce a functional gene product. 1. Students conduct investigations. Gene products are either proteins or noncoding RNAs, such 2. Students develop conclusions based on evidence. as tRNA and rRNA, which play essential roles in protein syn thesis, but do not code for proteins. Gene expression is regu Materials And AdvanCE Preparation lated throughout the lifespan of an individual cell to control the cell’s functions, such as its metabolic activity. Gene For the teacher expression plays a critical role in the morphological changes transparency of Student Sheet 2.3, “Genetics Case Study that take place in a developing embryo and fetus and in the Comparison” differentiation of stem cells to form specialized cells. -

Cell-Type-Specific Expression Quantitative Trait Loci Associated with Alzheimer Disease in Blood and Brain Tissue
Patel et al. Translational Psychiatry (2021) 11:250 https://doi.org/10.1038/s41398-021-01373-z Translational Psychiatry ARTICLE Open Access Cell-type-specific expression quantitative trait loci associated with Alzheimer disease in blood and brain tissue Devanshi Patel1,2, Xiaoling Zhang2,3,JohnJ.Farrell2, Jaeyoon Chung 2,ThorD.Stein4,5,6, Kathryn L. Lunetta 3 and Lindsay A. Farrer 1,2,3,7,8 Abstract Because regulation of gene expression is heritable and context-dependent, we investigated AD-related gene expression patterns in cell types in blood and brain. Cis-expression quantitative trait locus (eQTL) mapping was performed genome-wide in blood from 5257 Framingham Heart Study (FHS) participants and in brain donated by 475 Religious Orders Study/Memory & Aging Project (ROSMAP) participants. The association of gene expression with genotypes for all cis SNPs within 1 Mb of genes was evaluated using linear regression models for unrelated subjects and linear-mixed models for related subjects. Cell-type-specific eQTL (ct-eQTL) models included an interaction term for the expression of “proxy” genes that discriminate particular cell type. Ct-eQTL analysis identified 11,649 and 2533 additional significant gene-SNP eQTL pairs in brain and blood, respectively, that were not detected in generic eQTL analysis. Of note, 386 unique target eGenes of significant eQTLs shared between blood and brain were enriched in apoptosis and Wnt signaling pathways. Five of these shared genes are established AD loci. The potential importance and relevance to AD of significant results in myeloid cell types is supported by the observation that a large portion of fi 1234567890():,; 1234567890():,; 1234567890():,; 1234567890():,; GWS ct-eQTLs map within 1 Mb of established AD loci and 58% (23/40) of the most signi cant eGenes in these eQTLs have previously been implicated in AD.