Single-Cell Gene Expression Variation As a Cell-Type Specific Trait: a Study of Mammalian Gene Expression Using Single-Cell RNA Sequencing
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

FINALIST DIRECTORY VIRTUAL REGENERON ISEF 2020 Animal Sciences
FINALIST DIRECTORY VIRTUAL REGENERON ISEF 2020 Animal Sciences ANIM001 Dispersal and Behavior Patterns between ANIM010 The Study of Anasa tristis Elimination Using Dispersing Wolves and Pack Wolves in Northern Household Products Minnesota Carter McGaha, 15, Freshman, Vici Public Schools, Marcy Ferriere, 18, Senior, Cloquet, Senior High Vici, OK School, Cloquet, MN ANIM011 The Ketogenic Diet Ameliorates the Effects of ANIM002 Antsel and Gretal Caffeine in Seizure Susceptible Drosophila Avneesh Saravanapavan, 14, Freshman, West Port melanogaster High School, Ocala, FL Katherine St George, 17, Senior, John F. Kennedy High School, Bellmore, NY ANIM003 Year Three: Evaluating the Effects of Bifidobacterium infantis Compared with ANIM012 Development and Application of Attractants and Fumagillin on the Honeybee Gut Parasite Controlled-release Microcapsules for the Nosema ceranae and Overall Gut Microbiota Control of an Important Economic Pest: Flower # Varun Madan, 15, Sophomore, Lake Highland Thrips, Frankliniella intonsa Preparatory School, Orlando, FL Chunyi Wei, 16, Sophomore, The Affiliated High School of Fujian Normal University, Fuzhou, ANIM004T Using Protease-activated Receptors (PARs) in Fujian, China Caenorhabditis elegans as a Potential Therapeutic Agent for Inflammatory Diseases ANIM013 The Impacts of Brandt's Voles (Lasiopodomys Swetha Velayutham, 15, Sophomore, brandtii) on the Growth of Plantations Vyshnavi Poruri, 15, Sophomore, Surrounding their Patched Burrow Units Plano East, Senior High School, Plano, TX Meiqi Sun, 18, Senior, -

Single-Cell Transcriptome Analysis As a Promising Tool to Study Pluripotent Stem Cell Reprogramming
International Journal of Molecular Sciences Review Single-Cell Transcriptome Analysis as a Promising Tool to Study Pluripotent Stem Cell Reprogramming Hyun Kyu Kim †, Tae Won Ha † and Man Ryul Lee * Soonchunhyang Institute of Medi-Bio Science (SIMS), Soon Chun Hyang University, Cheonan 31151, Korea; [email protected] (H.K.K.); [email protected] (T.W.H.) * Correspondence: [email protected]; Tel.: +81-41-413-5013 † These authors equally contributed to this work. Abstract: Cells are the basic units of all organisms and are involved in all vital activities, such as proliferation, differentiation, senescence, and apoptosis. A human body consists of more than 30 trillion cells generated through repeated division and differentiation from a single-cell fertilized egg in a highly organized programmatic fashion. Since the recent formation of the Human Cell Atlas consortium, establishing the Human Cell Atlas at the single-cell level has been an ongoing activity with the goal of understanding the mechanisms underlying diseases and vital cellular activities at the level of the single cell. In particular, transcriptome analysis of embryonic stem cells at the single-cell level is of great importance, as these cells are responsible for determining cell fate. Here, we review single-cell analysis techniques that have been actively used in recent years, introduce the single-cell analysis studies currently in progress in pluripotent stem cells and reprogramming, and forecast future studies. Keywords: single-cell mRNA sequencing; pluripotent stem cell; somatic cell reprogramming; in- duced pluripotent stem cell; heterogeneity Citation: Kim, H.K.; Ha, T.W.; Lee, M.R. Single-Cell Transcriptome Analysis as a Promising Tool to Study Pluripotent Stem Cell Reprogramming. -

Introduction to the Cell Cell History Cell Structures and Functions
Introduction to the cell cell history cell structures and functions CK-12 Foundation December 16, 2009 CK-12 Foundation is a non-profit organization with a mission to reduce the cost of textbook materials for the K-12 market both in the U.S. and worldwide. Using an open-content, web-based collaborative model termed the “FlexBook,” CK-12 intends to pioneer the generation and distribution of high quality educational content that will serve both as core text as well as provide an adaptive environment for learning. Copyright ©2009 CK-12 Foundation This work is licensed under the Creative Commons Attribution-Share Alike 3.0 United States License. To view a copy of this license, visit http://creativecommons.org/licenses/by-sa/3.0/us/ or send a letter to Creative Commons, 171 Second Street, Suite 300, San Francisco, California, 94105, USA. Contents 1 Cell structure and function dec 16 5 1.1 Lesson 3.1: Introduction to Cells .................................. 5 3 www.ck12.org www.ck12.org 4 Chapter 1 Cell structure and function dec 16 1.1 Lesson 3.1: Introduction to Cells Lesson Objectives • Identify the scientists that first observed cells. • Outline the importance of microscopes in the discovery of cells. • Summarize what the cell theory proposes. • Identify the limitations on cell size. • Identify the four parts common to all cells. • Compare prokaryotic and eukaryotic cells. Introduction Knowing the make up of cells and how cells work is necessary to all of the biological sciences. Learning about the similarities and differences between cell types is particularly important to the fields of cell biology and molecular biology. -

PBMC Fixation and Processing for Chromium Single-Cell RNA Sequencing
bioRxiv preprint doi: https://doi.org/10.1101/315267; this version posted May 5, 2018. The copyright holder has placed this preprint (which was not certified by peer review) in the Public Domain. It is no longer restricted by copyright. Anyone can legally share, reuse, remix, or adapt this material for any purpose without crediting the original authors. PBMC Fixation and Processing for Chromium Single-Cell RNA Sequencing Jinguo Chen1, Foo Cheung1, Rongye Shi1,2, Huizhi Zhou1,2, Wenrui Lu1,3, CHI Consortium1 1Center for Human Immunology, Autoimmunity and Inflammation (CHI), National Institutes of Health 2Department of Transfusion Medicine, Clinical Center, National Institutes of Health 3Fujian Provincial Hospital, Fuzhou, China CHI Consortium (Julián Candia1, Yuri Kotliarov1, Katie R. Stagliano1 and John S. Tsang1,2) 1 CHI 2 Systems Genomics and Bioinformatics Unit, NIAID Abstract Background: Interest in single-cell transcriptomic analysis is growing rapidly, especially for profiling rare or heterogeneous populations of cells. In almost all reported works, investigators have used live cells which introduces cell stress during preparation and hinders complex study designs. Recent studies have indicated that cells fixed by denaturing fixative can be used in single-cell sequencing. But they did not work with most primary cells including immune cells. Methods: The methanol-fixation and new processing method was introduced to preserve PBMCs for single-cell RNA sequencing (scRNA-Seq) analysis on 10X Chromium platform. Results: When methanol fixation protocol was broken up into three steps: fixation, storage and rehydration, we found that PBMC RNA was degraded during rehydration with PBS, not at cell fixation and up to three-month storage steps. -
Creating a Census of Human Cells
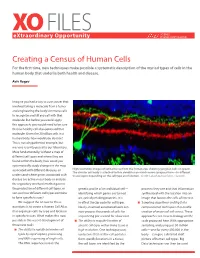
XO FILES SCIENCE eXtraordinary Opportunity PHILANTHROPY ALLIANCE Creating a Census of Human Cells For the first time, new techniques make possible a systematic description of the myriad types of cells in the human body that underlie both health and disease. Aviv Regev Imagine you had a way to cure cancer that involved taking a molecule from a tumor and engineering the body’s immune cells to recognize and kill any cell with that molecule. But before you could apply this approach, you would need to be sure that no healthy cell also expressed that molecule. Given the 20 trillion cells in a human body, how would you do that? This is not a hypothetical example, but was one recently posed to our laboratory. More fundamentally, without a map of different cell types and where they are found within the body, how could you systematically study changes in the map associated with different diseases, or High resolution images of retinal tissue from the human eye, showing ganglion cells (in green). The circular cell body is attached to thin dendrites on which nerve synapses form—in different understand where genes associated with tissue layers depending on the cell type and function. Credit: Lab of Joshua Sanes, Harvard. disease are active in our body or analyze the regulatory mechanisms that govern the production of different cell types, or genetic profile of an individual cell— proteins they use and that information sort out how different cell types combine identifying which genes are turned synthesized with the location into an to form specific tissues? on, actively making proteins. -

Robustness and Timing of Cellular Differentiation Through Population-Based Symmetry Breaking
AR TI CLE Robustness AND TIMING OF CELLULAR DIFFERENTIATION THROUGH population-based SYMMETRY BREAKING Angel StanoeV1 | Christian Schröter1 | Aneta KOSESKA1∗ 1Department OF Systemic Cell Biology, Max Planck Institute OF Molecular PhYSIOLOGY, Although COORDINATED BEHAVIOR OF MULTICELLULAR ORGANISMS Dortmund, GermanY RELIES ON INTERCELLULAR communication, THE DYNAMICAL mech- Correspondence ANISM THAT UNDERLIES SYMMETRY BREAKING IN HOMOGENEOUS Email: populations, THEREBY GIVING RISE TO HETEROGENEOUS CELL TYPES ∗[email protected] REMAINS UNCLEAR. In CONTRAST TO THE PREVALENT cell-intrinsic VIEW OF DIFFERENTIATION WHERE MULTISTABILITY ON THE SINGLE CELL LEVEL IS A NECESSARY pre-requisite, WE PROPOSE THAT het- EROGENEOUS CELLULAR IDENTITIES EMERGE AND ARE MAINTAINED COOPERATIVELY ON A POPULATION LEVel. Identifying A GENERIC SYMMETRY BREAKING mechanism, WE DEMONSTRATE THAT ROBUST PROPORTIONS BETWEEN DIFFERENTIATED CELL TYPES ARE INTRINSIC FEATURES OF THIS INHOMOGENEOUS STEADY STATE solution. Crit- ICAL ORGANIZATION IN THE VICINITY OF THE SYMMETRY BREAKING BIFURCATION ON THE OTHER hand, SUGGESTS THAT TIMING OF cel- LULAR DIFFERENTIATION EMERGES FOR GROWING POPULATIONS IN A self-organized MANNER. Setting A CLEAR DISTINCTION BETWEEN pre-eXISTING ASYMMETRIES AND SYMMETRY BREAKING EVents, WE THUS DEMONSTRATE THAT EMERGENT SYMMETRY breaking, IN CONJUNCTION WITH ORGANIZATION NEAR CRITICALITY NECESSARILY LEADS TO ROBUSTNESS AND ACCURACY DURING DEVelopment. KEYWORDS SYMMETRY breaking; differentiation; INHOMOGENEOUS STEADY STATE Abbreviations: -

A Concise Review on the Classification and Nomenclature of Stem Cells Kök Hücrelerinin S›N›Fland›R›Lmas› Ve Isimlendirilmesine Iliflkin K›Sa Bir Derleme
Review 57 A concise review on the classification and nomenclature of stem cells Kök hücrelerinin s›n›fland›r›lmas› ve isimlendirilmesine iliflkin k›sa bir derleme Alp Can Ankara University Medical School, Department of Histology and Embryology, Ankara, Turkey Abstract Stem cell biology and regenerative medicine is a relatively young field. However, in recent years there has been a tremen- dous interest in stem cells possibly due to their therapeutic potential in disease states. As a classical definition, a stem cell is an undifferentiated cell that can produce daughter cells that can either remain a stem cell in a process called self-renew- al, or commit to a specific cell type via the initiation of a differentiation pathway leading to the production of mature progeny cells. Despite this acknowledged definition, the classification of stem cells has been a perplexing notion that may often raise misconception even among stem cell biologists. Therefore, the aim of this brief review is to give a conceptual approach to classifying the stem cells beginning from the early morula stage totipotent embryonic stem cells to the unipotent tissue-resident adult stem cells, also called tissue-specific stem cells. (Turk J Hematol 2008; 25: 57-9) Key words: Stem cells, embryonic stem cells, tissue-specific stem cells, classification, progeny. Özet Kök hücresi biyolojisi ve onar›msal t›p görece yeni alanlard›r. Buna karfl›n, son y›llarda çeflitli hastal›klarda tedavi amac›yla kullan›labilme potansiyelleri nedeniyle kök hücrelerine ola¤anüstü bir ilgi art›fl› vard›r. Klasik tan›m›yla kök hücresi, kendini yenileme ad› verilen mekanizmayla farkl›laflmadan kendini ço¤altan veya bir dizi farkl›laflma aflamas›ndan geçerek olgun hücrelere dönüflebilen hücrelerdir. -

Shapes TCR Usage in Celiac Disease Posttranslational Modification of Gluten
Posttranslational Modification of Gluten Shapes TCR Usage in Celiac Disease Shuo-Wang Qiao, Melinda Ráki, Kristin S. Gunnarsen, Geir-Åge Løset, Knut E. A. Lundin, Inger Sandlie and This information is current as Ludvig M. Sollid of September 29, 2021. J Immunol 2011; 187:3064-3071; Prepublished online 17 August 2011; doi: 10.4049/jimmunol.1101526 http://www.jimmunol.org/content/187/6/3064 Downloaded from Supplementary http://www.jimmunol.org/content/suppl/2011/08/18/jimmunol.110152 Material 6.DC1 http://www.jimmunol.org/ References This article cites 31 articles, 12 of which you can access for free at: http://www.jimmunol.org/content/187/6/3064.full#ref-list-1 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision by guest on September 29, 2021 • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2011 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology Posttranslational Modification of Gluten Shapes TCR Usage in Celiac Disease Shuo-Wang Qiao,* Melinda Ra´ki,†,1 Kristin S. -

Single-Cell Sequencing for Precise Cancer Research: Progress and Prospects Xiaoyan Zhang1, Sadie L
Published OnlineFirst March 3, 2016; DOI: 10.1158/0008-5472.CAN-15-1907 Cancer Review Research Single-Cell Sequencing for Precise Cancer Research: Progress and Prospects Xiaoyan Zhang1, Sadie L. Marjani2, Zhaoyang Hu1, Sherman M. Weissman3, Xinghua Pan1,3,4, and Shixiu Wu1 Abstract Advances in genomic technology have enabled the faithful could potentially improve the early detection and monitoring detection and measurement of mutations and the gene expres- of rare cancer cells, such as circulating tumor cells and dissem- sion profile of cancer cells at the single-cell level. Recently, inated tumor cells, and promote the development of person- several single-cell sequencing methods have been developed alized and highly precise cancer therapy. Here, we discuss that permit the comprehensive and precise analysis of the the current methods for single cancer-cell sequencing, with a cancer-cell genome, transcriptome, and epigenome. The use of strong focus on those practically used or potentially valuable these methods to analyze cancer cells has led to a series of in cancer research, including single-cell isolation, whole unanticipated discoveries, such as the high heterogeneity and genome and transcriptome amplification, epigenome profiling, stochastic changes in cancer-cell populations, the new driver multi-dimensional sequencing, and next-generation sequenc- mutations and the complicated clonal evolution mechanisms, ing and analysis. We also examine the current applications, and the novel identification of biomarkers of variant tumors. challenges, and prospects of single cancer-cell sequencing. These methods and the knowledge gained from their utilization Cancer Res; 76(6); 1–8. Ó2016 AACR. Introduction disease progression (4). However, the molecular profiling data, especially heterogeneity analysis, of these important cancer-cell Cancer is a significant cause of mortality worldwide. -

Polish Roadmap for Research Infrastructures Polish Roadmap for Research Infrastructures
Polish Roadmap for Research Infrastructures Polish Roadmap for Research Infrastructures Preface Research plays a key role in the development of soci- ety and civilization. Through research we are able to ef- fectively address new challenges such as quality of life, civilization diseases, demographic changes, sustainable development and environmental protection, as well as energy security. Without top-level research it is impos- sible not only to successfully face the above mentioned and other challenges but also to understand the dy- Wojciech Murdzek namically changing world. Minister of Science and Higher Education There are two elements of vital importance for research excellence – human capital and state-of-the-art re- search infrastructures. The latter is doubly important owing to its essential role in training future scientific and technical staff. Major strategic research infrastruc- tures attract world-class scientists and innovative com- panies which enables economic growth as well as en- hance social capital. Excellent laboratories applying the highest standards of research and education are there- fore necessary for each country to develop. The Polish Roadmap for Research Infrastructures is a tool created to facilitate the development of these laboratories. It comprises seventy of the most ambitious research infrastructure projects of great socio-econom- ic significance, chosen through a transparent process carried out in accordance with international standards. The Roadmap is to become the basis for the investment policy of the Ministry of Science and Higher Education in the coming years, however it is worth underlining that placing a research infrastructure project on the Roadmap does not automatically imply the financial commitment of the Ministry. -

Exploring Viral Infection Using Single-Cell Sequencing
Virus Research 239 (2017) 55–68 Contents lists available at ScienceDirect Virus Research j ournal homepage: www.elsevier.com/locate/virusres Review Exploring viral infection using single-cell sequencing a,∗ b c a Sylvie Rato , Monica Golumbeanu , Amalio Telenti , Angela Ciuffi a Institute of Microbiology, University Hospital Center and University of Lausanne, Bugnon 48, CH-1011 Lausanne, Switzerland b Department of Biosystems Science and Engineering, ETH Zürich and SIB Swiss Institute of Bioinformatics, CH-4058 Basel, Switzerland c The J. Craig Venter Institute, 4120 Capricorn Lane, La Jolla, CA 92037, USA a r t i c l e i n f o a b s t r a c t Article history: Single-cell sequencing (SCS) has emerged as a valuable tool to study cellular heterogeneity in diverse Received 7 June 2016 fields, including virology. By studying the viral and cellular genome and/or transcriptome, the dynamics Received in revised form 21 October 2016 of viral infection can be investigated at single cell level. Most studies have explored the impact of cell-to- Accepted 24 October 2016 cell variation on the viral life cycle from the point of view of the virus, by analyzing viral sequences, and Available online 2 November 2016 from the point of view of the cell, mainly by analyzing the cellular host transcriptome. In this review, we will focus on recent studies that use single-cell sequencing to explore viral diversity and cell variability Keywords: in response to viral replication. Single-cell Heterogeneity © 2016 The Authors. Published by Elsevier B.V. This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/4.0/). -

An Introduction to Stem Cell Biology
An Introduction to Stem Cell Biology Michael L. Shelanski, MD,PhD Professor of Pathology and Cell Biology Columbia University Figures adapted from ISSCR. Presentations of Drs. Martin Pera (Monash University), Dr.Susan Kadereit, Children’s Hospital, Boston and Dr. Catherine Verfaillie, University of Minnesota Science 1999, 283: 534-537 PNAS 1999, 96: 14482-14486 Turning Blood into Brain: Cells Bearing Neuronal Antigens Generated in Vitro from Bone Marrow Science 2000, 290:1779-1782 From Marrow to Brain: Expression of Neuronal Phenotypes in Adult Mice Mezey, E., Chandross, K.J., Harta, G., Maki, R.A., McKercher, S.R. Science 2000, 290:1775-1779 Brazelton, T.R., Rossi, F.M., Keshet, G.I., Blau, H.M. Nature 2001, 410:701-705 Nat Med 2000, 11: 1229-1234 Stem Cell FAQs Do you need to get one from an egg? Must you sacrifice an Embryo? What is an ES cell? What about adult stem cells or cord blood stem cells Why can’t this work be done in animals? Are “cures” on the horizon? Will this lead to human cloning – human spare parts factories? Are we going to make a Frankenstein? What is a stem cell? A primitive cell which can either self renew (reproduce itself) or give rise to more specialised cell types The stem cell is the ancestor at the top of the family tree of related cell types. One blood stem cell gives rise to red cells, white cells and platelets Stem Cells Vary in their Developmental capacity A multipotent cell can give rise to several types of mature cell A pluripotent cell can give rise to all types of adult tissue cells plus extraembryonic tissue: cells which support embryonic development A totipotent cell can give rise to a new individual given appropriate maternal support The Fertilized Egg The “Ultimate” Stem Cell – the Newly Fertilized Egg (one Cell) will give rise to all the cells and tissues of the adult animal.