FUT9-Driven Programming of Colon Cancer Cells Towards a Stem Cell-Like State
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Systematic Review on the Implications of O-Linked Glycan Branching and Truncating Enzymes on Cancer Progression and Metastasis
cells Review A Systematic Review on the Implications of O-linked Glycan Branching and Truncating Enzymes on Cancer Progression and Metastasis 1, 1, 1 1,2,3, Rohitesh Gupta y, Frank Leon y, Sanchita Rauth , Surinder K. Batra * and Moorthy P. Ponnusamy 1,2,* 1 Department of Biochemistry and Molecular Biology, University of Nebraska Medical Center, Omaha, NE 68105, USA; [email protected] (R.G.); [email protected] (F.L.); [email protected] (S.R.) 2 Fred and Pamela Buffett Cancer Center, Eppley Institute for Research in Cancer and Allied Diseases, University of Nebraska Medical Center, Omaha, NE 681980-5900, USA 3 Department of Pathology and Microbiology, UNMC, Omaha, NE 68198-5900, USA * Correspondence: [email protected] (S.K.B.); [email protected] (M.P.P.); Tel.: +402-559-5455 (S.K.B.); +402-559-1170 (M.P.P.); Fax: +402-559-6650 (S.K.B. & M.P.P.) Equal contribution. y Received: 21 January 2020; Accepted: 12 February 2020; Published: 14 February 2020 Abstract: Glycosylation is the most commonly occurring post-translational modifications, and is believed to modify over 50% of all proteins. The process of glycan modification is directed by different glycosyltransferases, depending on the cell in which it is expressed. These small carbohydrate molecules consist of multiple glycan families that facilitate cell–cell interactions, protein interactions, and downstream signaling. An alteration of several types of O-glycan core structures have been implicated in multiple cancers, largely due to differential glycosyltransferase expression or activity. Consequently, aberrant O-linked glycosylation has been extensively demonstrated to affect biological function and protein integrity that directly result in cancer growth and progression of several diseases. -

Characterization of Genomic Copy Number Variation in Mus Musculus Associated with the Germline of Inbred and Wild Mouse Populations, Normal Development, and Cancer
Western University Scholarship@Western Electronic Thesis and Dissertation Repository 4-18-2019 2:00 PM Characterization of genomic copy number variation in Mus musculus associated with the germline of inbred and wild mouse populations, normal development, and cancer Maja Milojevic The University of Western Ontario Supervisor Hill, Kathleen A. The University of Western Ontario Graduate Program in Biology A thesis submitted in partial fulfillment of the equirr ements for the degree in Doctor of Philosophy © Maja Milojevic 2019 Follow this and additional works at: https://ir.lib.uwo.ca/etd Part of the Genetics and Genomics Commons Recommended Citation Milojevic, Maja, "Characterization of genomic copy number variation in Mus musculus associated with the germline of inbred and wild mouse populations, normal development, and cancer" (2019). Electronic Thesis and Dissertation Repository. 6146. https://ir.lib.uwo.ca/etd/6146 This Dissertation/Thesis is brought to you for free and open access by Scholarship@Western. It has been accepted for inclusion in Electronic Thesis and Dissertation Repository by an authorized administrator of Scholarship@Western. For more information, please contact [email protected]. Abstract Mus musculus is a human commensal species and an important model of human development and disease with a need for approaches to determine the contribution of copy number variants (CNVs) to genetic variation in laboratory and wild mice, and arising with normal mouse development and disease. Here, the Mouse Diversity Genotyping array (MDGA)-approach to CNV detection is developed to characterize CNV differences between laboratory and wild mice, between multiple normal tissues of the same mouse, and between primary mammary gland tumours and metastatic lung tissue. -

Introduction to the Cell Cell History Cell Structures and Functions
Introduction to the cell cell history cell structures and functions CK-12 Foundation December 16, 2009 CK-12 Foundation is a non-profit organization with a mission to reduce the cost of textbook materials for the K-12 market both in the U.S. and worldwide. Using an open-content, web-based collaborative model termed the “FlexBook,” CK-12 intends to pioneer the generation and distribution of high quality educational content that will serve both as core text as well as provide an adaptive environment for learning. Copyright ©2009 CK-12 Foundation This work is licensed under the Creative Commons Attribution-Share Alike 3.0 United States License. To view a copy of this license, visit http://creativecommons.org/licenses/by-sa/3.0/us/ or send a letter to Creative Commons, 171 Second Street, Suite 300, San Francisco, California, 94105, USA. Contents 1 Cell structure and function dec 16 5 1.1 Lesson 3.1: Introduction to Cells .................................. 5 3 www.ck12.org www.ck12.org 4 Chapter 1 Cell structure and function dec 16 1.1 Lesson 3.1: Introduction to Cells Lesson Objectives • Identify the scientists that first observed cells. • Outline the importance of microscopes in the discovery of cells. • Summarize what the cell theory proposes. • Identify the limitations on cell size. • Identify the four parts common to all cells. • Compare prokaryotic and eukaryotic cells. Introduction Knowing the make up of cells and how cells work is necessary to all of the biological sciences. Learning about the similarities and differences between cell types is particularly important to the fields of cell biology and molecular biology. -

Patient-Based Cross-Platform Comparison of Oligonucleotide Microarray Expression Profiles
Laboratory Investigation (2005) 85, 1024–1039 & 2005 USCAP, Inc All rights reserved 0023-6837/05 $30.00 www.laboratoryinvestigation.org Patient-based cross-platform comparison of oligonucleotide microarray expression profiles Joerg Schlingemann1,*, Negusse Habtemichael2,*, Carina Ittrich3, Grischa Toedt1, Heidi Kramer1, Markus Hambek4, Rainald Knecht4, Peter Lichter1, Roland Stauber2 and Meinhard Hahn1 1Division of Molecular Genetics, Deutsches Krebsforschungszentrum, Heidelberg, Germany; 2Chemotherapeutisches Forschungsinstitut Georg-Speyer-Haus, Frankfurt am Main, Germany; 3Central Unit Biostatistics, Deutsches Krebsforschungszentrum, Heidelberg, Germany and 4Department of Otorhinolaryngology, Universita¨tsklinik, Johann-Wolfgang-Goethe-Universita¨t Frankfurt, Frankfurt, Germany The comparison of gene expression measurements obtained with different technical approaches is of substantial interest in order to clarify whether interplatform differences may conceal biologically significant information. To address this concern, we analyzed gene expression in a set of head and neck squamous cell carcinoma patients, using both spotted oligonucleotide microarrays made from a large collection of 70-mer probes and commercial arrays produced by in situ synthesis of sets of multiple 25-mer oligonucleotides per gene. Expression measurements were compared for 4425 genes represented on both platforms, which revealed strong correlations between the corresponding data sets. Of note, a global tendency towards smaller absolute ratios was observed when -
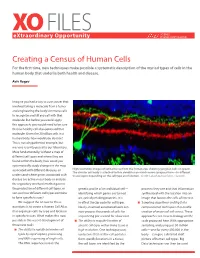
Creating a Census of Human Cells
XO FILES SCIENCE eXtraordinary Opportunity PHILANTHROPY ALLIANCE Creating a Census of Human Cells For the first time, new techniques make possible a systematic description of the myriad types of cells in the human body that underlie both health and disease. Aviv Regev Imagine you had a way to cure cancer that involved taking a molecule from a tumor and engineering the body’s immune cells to recognize and kill any cell with that molecule. But before you could apply this approach, you would need to be sure that no healthy cell also expressed that molecule. Given the 20 trillion cells in a human body, how would you do that? This is not a hypothetical example, but was one recently posed to our laboratory. More fundamentally, without a map of different cell types and where they are found within the body, how could you systematically study changes in the map associated with different diseases, or High resolution images of retinal tissue from the human eye, showing ganglion cells (in green). The circular cell body is attached to thin dendrites on which nerve synapses form—in different understand where genes associated with tissue layers depending on the cell type and function. Credit: Lab of Joshua Sanes, Harvard. disease are active in our body or analyze the regulatory mechanisms that govern the production of different cell types, or genetic profile of an individual cell— proteins they use and that information sort out how different cell types combine identifying which genes are turned synthesized with the location into an to form specific tissues? on, actively making proteins. -

Investigation of the Underlying Hub Genes and Molexular Pathogensis in Gastric Cancer by Integrated Bioinformatic Analyses
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.20.423656; this version posted December 22, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Investigation of the underlying hub genes and molexular pathogensis in gastric cancer by integrated bioinformatic analyses Basavaraj Vastrad1, Chanabasayya Vastrad*2 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karanataka, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2020.12.20.423656; this version posted December 22, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract The high mortality rate of gastric cancer (GC) is in part due to the absence of initial disclosure of its biomarkers. The recognition of important genes associated in GC is therefore recommended to advance clinical prognosis, diagnosis and and treatment outcomes. The current investigation used the microarray dataset GSE113255 RNA seq data from the Gene Expression Omnibus database to diagnose differentially expressed genes (DEGs). Pathway and gene ontology enrichment analyses were performed, and a proteinprotein interaction network, modules, target genes - miRNA regulatory network and target genes - TF regulatory network were constructed and analyzed. Finally, validation of hub genes was performed. The 1008 DEGs identified consisted of 505 up regulated genes and 503 down regulated genes. -

Robustness and Timing of Cellular Differentiation Through Population-Based Symmetry Breaking
AR TI CLE Robustness AND TIMING OF CELLULAR DIFFERENTIATION THROUGH population-based SYMMETRY BREAKING Angel StanoeV1 | Christian Schröter1 | Aneta KOSESKA1∗ 1Department OF Systemic Cell Biology, Max Planck Institute OF Molecular PhYSIOLOGY, Although COORDINATED BEHAVIOR OF MULTICELLULAR ORGANISMS Dortmund, GermanY RELIES ON INTERCELLULAR communication, THE DYNAMICAL mech- Correspondence ANISM THAT UNDERLIES SYMMETRY BREAKING IN HOMOGENEOUS Email: populations, THEREBY GIVING RISE TO HETEROGENEOUS CELL TYPES ∗[email protected] REMAINS UNCLEAR. In CONTRAST TO THE PREVALENT cell-intrinsic VIEW OF DIFFERENTIATION WHERE MULTISTABILITY ON THE SINGLE CELL LEVEL IS A NECESSARY pre-requisite, WE PROPOSE THAT het- EROGENEOUS CELLULAR IDENTITIES EMERGE AND ARE MAINTAINED COOPERATIVELY ON A POPULATION LEVel. Identifying A GENERIC SYMMETRY BREAKING mechanism, WE DEMONSTRATE THAT ROBUST PROPORTIONS BETWEEN DIFFERENTIATED CELL TYPES ARE INTRINSIC FEATURES OF THIS INHOMOGENEOUS STEADY STATE solution. Crit- ICAL ORGANIZATION IN THE VICINITY OF THE SYMMETRY BREAKING BIFURCATION ON THE OTHER hand, SUGGESTS THAT TIMING OF cel- LULAR DIFFERENTIATION EMERGES FOR GROWING POPULATIONS IN A self-organized MANNER. Setting A CLEAR DISTINCTION BETWEEN pre-eXISTING ASYMMETRIES AND SYMMETRY BREAKING EVents, WE THUS DEMONSTRATE THAT EMERGENT SYMMETRY breaking, IN CONJUNCTION WITH ORGANIZATION NEAR CRITICALITY NECESSARILY LEADS TO ROBUSTNESS AND ACCURACY DURING DEVelopment. KEYWORDS SYMMETRY breaking; differentiation; INHOMOGENEOUS STEADY STATE Abbreviations: -

Detecting Substrate Glycans of Fucosyltransferases on Glycoproteins with Fluorescent Fucose
bioRxiv preprint doi: https://doi.org/10.1101/2020.01.28.919860; this version posted January 29, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Detecting substrate glycans of fucosyltransferases on glycoproteins with fluorescent fucose Key words: Fucose/Fucosylation/fucosyltransferase/core-fucose/glycosylation Supplementary Data Included: Supplemental Fig.1 to Fig. 2 Zhengliang L Wu1*, Mark Whitaker, Anthony D Person1, Vassili Kalabokis1 1Bio-techne, R&D Systems, Inc. 614 McKinley Place N.E. Minneapolis, MN, 55413, USA *Correspondence: Phone: 612-656-4544. Email: [email protected], bioRxiv preprint doi: https://doi.org/10.1101/2020.01.28.919860; this version posted January 29, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Abstract Like sialylation, fucose usually locates at the non-reducing ends of various glycans on glycoproteins and constitutes important glycan epitopes. Detecting the substrate glycans of fucosyltransferases is important for understanding how these glycan epitopes are regulated in response to different growth conditions and external stimuli. Here we report the detection of these glycans via enzymatic incorporation of fluorescent tagged fucose using fucosyltransferases including FUT2, FUT6, FUT7, and FUT8 and FUT9. More specifically, we describe the detection of substrate glycans of FUT8 and FUT9 on therapeutic antibodies and the detection of high mannose glycans on glycoproteins by enzymatic conversion of high mannose glycans to the substrate glycans of FUT8. -

A Concise Review on the Classification and Nomenclature of Stem Cells Kök Hücrelerinin S›N›Fland›R›Lmas› Ve Isimlendirilmesine Iliflkin K›Sa Bir Derleme
Review 57 A concise review on the classification and nomenclature of stem cells Kök hücrelerinin s›n›fland›r›lmas› ve isimlendirilmesine iliflkin k›sa bir derleme Alp Can Ankara University Medical School, Department of Histology and Embryology, Ankara, Turkey Abstract Stem cell biology and regenerative medicine is a relatively young field. However, in recent years there has been a tremen- dous interest in stem cells possibly due to their therapeutic potential in disease states. As a classical definition, a stem cell is an undifferentiated cell that can produce daughter cells that can either remain a stem cell in a process called self-renew- al, or commit to a specific cell type via the initiation of a differentiation pathway leading to the production of mature progeny cells. Despite this acknowledged definition, the classification of stem cells has been a perplexing notion that may often raise misconception even among stem cell biologists. Therefore, the aim of this brief review is to give a conceptual approach to classifying the stem cells beginning from the early morula stage totipotent embryonic stem cells to the unipotent tissue-resident adult stem cells, also called tissue-specific stem cells. (Turk J Hematol 2008; 25: 57-9) Key words: Stem cells, embryonic stem cells, tissue-specific stem cells, classification, progeny. Özet Kök hücresi biyolojisi ve onar›msal t›p görece yeni alanlard›r. Buna karfl›n, son y›llarda çeflitli hastal›klarda tedavi amac›yla kullan›labilme potansiyelleri nedeniyle kök hücrelerine ola¤anüstü bir ilgi art›fl› vard›r. Klasik tan›m›yla kök hücresi, kendini yenileme ad› verilen mekanizmayla farkl›laflmadan kendini ço¤altan veya bir dizi farkl›laflma aflamas›ndan geçerek olgun hücrelere dönüflebilen hücrelerdir. -

Reciprocal Monoallelic Expression of ASAR Lncrna Genes Controls Replication Timing of Human Chromosome 6
Downloaded from rnajournal.cshlp.org on October 5, 2021 - Published by Cold Spring Harbor Laboratory Press 1 Reciprocal monoallelic expression of ASAR lncRNA genes 2 controls replication timing of human chromosome 6. 3 4 Michael Heskett1, Leslie G. Smith2, Paul Spellman1, and Mathew J. Thayer2* 5 6 1Department of Chemical Physiology and Biochemistry 7 Oregon Health & Science University 8 3181 S. W. Sam Jackson Park Road 9 Portland, Oregon 97239, USA 10 11 2Department of Chemical Physiology and Biochemistry 12 Oregon Health & Science University 13 3181 S. W. Sam Jackson Park Road 14 Portland, Oregon 97239, USA 15 16 *Corresponding author. 17 ORCID: 0000-0001-6483-1661 18 TEL: (503) 494-2447 19 FAX: (503) 494-7368 20 Email:[email protected] 21 22 Running Title: ASAR lincRNA genes on chromosome 6 23 Key Words: Replication timing, cis-acting element, non-coding RNA 1 Downloaded from rnajournal.cshlp.org on October 5, 2021 - Published by Cold Spring Harbor Laboratory Press 24 Abstract 25 DNA replication occurs on mammalian chromosomes in a cell-type distinctive temporal 26 order known as the replication timing program. We previously found that disruption of the 27 noncanonical lncRNA genes ASAR6 and ASAR15 results in delayed replication timing 28 and delayed mitotic chromosome condensation of human chromosomes 6 and 15, 29 respectively. ASAR6 and ASAR15 display random monoallelic expression, and display 30 asynchronous replication between alleles that is coordinated with other random 31 monoallelic genes on their respective chromosomes. Disruption of the expressed allele, 32 but not the silent allele, of ASAR6 leads to delayed replication, activation of the previously 33 silent alleles of linked monoallelic genes, and structural instability of human chromosome 34 6. -

B4GALT3 (B4GALT2) (NM 030587) Human Tagged ORF Clone Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for RC233988 B4GALT3 (B4GALT2) (NM_030587) Human Tagged ORF Clone Product data: Product Type: Expression Plasmids Product Name: B4GALT3 (B4GALT2) (NM_030587) Human Tagged ORF Clone Tag: Myc-DDK Symbol: B4GALT2 Synonyms: B4Gal-T2; B4Gal-T3; beta4Gal-T2 Vector: pCMV6-Entry (PS100001) E. coli Selection: Kanamycin (25 ug/mL) Cell Selection: Neomycin ORF Nucleotide >RC233988 representing NM_030587 Sequence: Red=Cloning site Blue=ORF Green=Tags(s) TTTTGTAATACGACTCACTATAGGGCGGCCGGGAATTCGTCGACTGGATCCGGTACCGAGGAGATCTGCC GCCGCGATCGCC ATGGCTGTGGAAGTCCAGGAGCAGTGGCCTTGTTTGCCAGCAGCCGGATGCCCGGGCCCACTGGGCGGGC CAGTGGCCGCCTGCGGGATGAGCAGACTGCTGGGGGGGACGCTGGAGCGCGTCTGCAAGGCTGTGCTCCT TCTCTGCCTGCTGCACTTCCTCGTGGCCGTCATCCTCTACTTTGACGTCTACGCCCAGCACCTGGCCTTC TTCAGCCGCTTCAGTGCCCGAGGCCCTGCCCATGCCCTCCACCCAGCTGCTAGCAGCAGCAGCAGCAGCA GCAACTGCTCCCGGCCCAACGCCACCGCCTCTAGCTCCGGGCTCCCTGAGGTCCCCAGTGCCCTGCCCGG TCCCACGGCTCCCACGCTGCCACCCTGTCCTGACTCGCCACCTGGTCTTGTGGGCAGACTGCTGATCGAG TTCACCTCACCCATGCCCCTGGAGCGGGTGCAGAGGGAGAACCCAGGCGTGCTCATGGGCGGCCGATACA CACCGCCCGACTGCACCCCAGCCCAGACGGTGGCGGTCATCATCCCCTTTAGACACCGGGAACACCACCT GCGCTACTGGCTCCACTATCTACACCCCATCTTGAGGCGGCAGCGGCTGCGCTACGGCGTCTATGTCATC AACCAGCATGGTGAGGACACCTTCAACCGGGCCAAGCTGCTTAACGTGGGCTTCCTAGAGGCGCTGAAGG AGGATGCCGCCTATGACTGCTTCATCTTCAGCGATGTGGACCTGGTCCCCATGGATGACCGCAACCTATA CCGCTGCGGCGACCAACCCCGCCACTTTGCCATTGCCATGGACAAGTTTGGCTTCCGGCTTCCCTATGCT -

Supplementary Data
Supplementary Fig. 1 A B Responder_Xenograft_ Responder_Xenograft_ NON- NON- Lu7336, Vehicle vs Lu7466, Vehicle vs Responder_Xenograft_ Responder_Xenograft_ Sagopilone, Welch- Sagopilone, Welch- Lu7187, Vehicle vs Lu7406, Vehicle vs Test: 638 Test: 600 Sagopilone, Welch- Sagopilone, Welch- Test: 468 Test: 482 Responder_Xenograft_ NON- Lu7860, Vehicle vs Responder_Xenograft_ Sagopilone, Welch - Lu7558, Vehicle vs Test: 605 Sagopilone, Welch- Test: 333 Supplementary Fig. 2 Supplementary Fig. 3 Supplementary Figure S1. Venn diagrams comparing probe sets regulated by Sagopilone treatment (10mg/kg for 24h) between individual models (Welsh Test ellipse p-value<0.001 or 5-fold change). A Sagopilone responder models, B Sagopilone non-responder models. Supplementary Figure S2. Pathway analysis of genes regulated by Sagopilone treatment in responder xenograft models 24h after Sagopilone treatment by GeneGo Metacore; the most significant pathway map representing cell cycle/spindle assembly and chromosome separation is shown, genes upregulated by Sagopilone treatment are marked with red thermometers. Supplementary Figure S3. GeneGo Metacore pathway analysis of genes differentially expressed between Sagopilone Responder and Non-Responder models displaying –log(p-Values) of most significant pathway maps. Supplementary Tables Supplementary Table 1. Response and activity in 22 non-small-cell lung cancer (NSCLC) xenograft models after treatment with Sagopilone and other cytotoxic agents commonly used in the management of NSCLC Tumor Model Response type