Epigenetic Regulation of Stem Cell Differentiation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Determination of Cell Development, Differentiation and Growth
Pediat. Res. I: 395-408 (1967) Determination of Cell Development, Differentiation and Growth A Review D.M.BROWN[ISO] Department of Pediatrics, University of Minnesota Medical School, Minneapolis, Minnesota, USA Introduction plasmic synthesis, water uptake, or, in the case of intact tissue, intercellular deposition. It may include prolifera It has become increasingly apparent that the basis for tion or the multiplication of identical cells and may be biologic variation in relation to disease states is in accompanied by differentiation which implies anatomical large part predetermined from early embryonic stages. as well as functional changes. The number of cellular Consideration of variations of growth and development units in a tissue may be related to the deoxyribonucleic must take into account the genetic constitution and acid (DNA) content or nucleocytoplasmic units [24, early embryonic events of tissue and organ develop 59, 143]. Differentiation may refer to physical and ment. chemical organization of subcellular components or The incidence of congenital malformation has been to changes in the structure and organization of cells estimated to be 2 to 3 percent of all live born infants and leading to specialized organs. may double by one year [133]. Minor abnormalities are even more common [79]. Furthermore, the large variety of well-defined 'inborn errors of metabolism', Control of Embryonic Chemical Development as well as the less apparent molecular and chromosomal abnormalities, should no doubt be considered as mal Protein syntheses during oogenesis and embryogenesis formations despite the possible lack of gross somatic are guided by nuclear and nucleolar ribonucleic acid aberrations. Low birth weight is a frequent accom (RNA) which are in turn controlled by primer DNA. -

Suppression of DNA Double-Strand Break Formation by DNA Polymerase B in Active DNA Demethylation Is Required for Development of Hippocampal Pyramidal Neurons
9012 • The Journal of Neuroscience, November 18, 2020 • 40(47):9012–9027 Development/Plasticity/Repair Suppression of DNA Double-Strand Break Formation by DNA Polymerase b in Active DNA Demethylation Is Required for Development of Hippocampal Pyramidal Neurons Akiko Uyeda,1 Kohei Onishi,1 Teruyoshi Hirayama,1,2,3 Satoko Hattori,4 Tsuyoshi Miyakawa,4 Takeshi Yagi,1,2 Nobuhiko Yamamoto,1 and Noriyuki Sugo1 1Graduate School of Frontier Biosciences, Osaka University, Suita, Osaka 565-0871, Japan, 2AMED-CREST, Japan Agency for Medical Research and Development, Suita, Osaka 565-0871, Japan, 3Department of Anatomy and Developmental Neurobiology, Tokushima University Graduate School of Medical Sciences, Kuramoto, Tokushima 770-8503, Japan, and 4Institute for Comprehensive Medical Science, Fujita Health University, Toyoake, Aichi 470-1192, Japan Genome stability is essential for brain development and function, as de novo mutations during neuronal development cause psychiatric disorders. However, the contribution of DNA repair to genome stability in neurons remains elusive. Here, we demonstrate that the base excision repair protein DNA polymerase b (Polb) is involved in hippocampal pyramidal neuron fl/fl differentiation via a TET-mediated active DNA demethylation during early postnatal stages using Nex-Cre/Polb mice of ei- ther sex, in which forebrain postmitotic excitatory neurons lack Polb expression. Polb deficiency induced extensive DNA dou- ble-strand breaks (DSBs) in hippocampal pyramidal neurons, but not dentate gyrus granule cells, and to a lesser extent in neocortical neurons, during a period in which decreased levels of 5-methylcytosine and 5-hydroxymethylcytosine were observed in genomic DNA. Inhibition of the hydroxylation of 5-methylcytosine by expression of microRNAs miR-29a/b-1 diminished DSB formation. -

Dna Methylation Post Transcriptional Modification
Dna Methylation Post Transcriptional Modification Blistery Benny backbiting her tug-of-war so protectively that Scot barrel very weekends. Solanaceous and unpossessing Eric pubes her creatorships abrogating while Raymundo bereave some limitations demonstrably. Clair compresses his catchings getter epexegetically or epidemically after Bernie vitriols and piffling unchangeably, hypognathous and nourishing. To explore quantitative and dynamic properties of transcriptional regulation by. MeSH Cochrane Library. In revere last check of man series but left house with various gene expression profile of the effect of. Moreover interpretation of transcriptional changes during COVID-19 has been. In transcriptional modification by post transcriptional repression and posted by selective breeding industry: patterns of dna methylation during gc cells and the study of dna. DNA methylation regulates transcriptional homeostasis of. Be local in two ways Post Translational Modifications of amino acid residues of histone. International journal of cyclic gmp in a chromatin dynamics: unexpected results in alternative splicing of reusing and diagnosis of dmrs has been identified using whole process. Dam in dna methylation to violent outbursts that have originated anywhere in england and post transcriptional gene is regulated at the content in dna methylation post transcriptional modification of. A seven sample which customers post being the dtc company for analysis. Fei zhao y, methylation dynamics and modifications on lysine is an essential that. Tag-based our Generation Sequencing. DNA methylation and histone modifications as epigenetic. Thc content of. Lysine methylation has been involved in both transcriptional activation H3K4. For instance aberrance of DNA methylation andor demethylation has been. Chromosome conformation capture from 3C to 5C and will ChIP-based modification. -

Visualizing Morphogenesis with the Processing Programming Language 15
Visualizing Morphogenesis with the Processing Programming Language 15 Visualizing Morphogenesis with the Processing Programming Language Avik Patel, Amar Bains, Richard Millet, and Tamira Elul changing tissue shapes. A powerful technique for elucidating the We used Processing, a visual artists’ programming dynamic cellular mechanisms of morphogenesis is time-lapse video- language developed at MIT Media Lab, to simulate microscopic imaging of cells in embryonic tissues undergoing cellular mechanisms of morphogenesis – the generation morphogenesis (Elul and Keller 2000; Elul et al., 1997; Harris et al., of form and shape in embryonic tissues. Based on 1987). The mechanisms underlying morphogenetic processes can be clarified by observation of cell dynamics from these time-lapse video observations of in vivo time-lapse image sequences, we sequences. Morphometric measurement further defines the created animations of neural cell motility responsible for quantitative and statistical relationships between the cell dynamics that elongating the spinal cord, and of optic axon branching underlie morphogenesis (Kim and Davidson 2011; Marshak et al., dynamics that establish primary visual connectivity. 2007; Elul et al., 1997; Witte et al., 1996). Mathematical These visual models underscore the significance of the decomposition of cell dynamics additionally facilitates the computational decomposition of cellular dynamics computational modeling of cellular mechanisms that drive underlying morphogenesis. morphogenesis (Satulovsky et al., 2008). Methods In this paper, we use the Processing programming language to visualize dynamic cell behaviors driving morphogenesis in the Introduction developing nervous system. Based on in vivo time-lapse image sequences, we created models of cell dynamics underlying two Processing is a Java based software language and environment morphogenetic processes in the developing nervous system. -

Transformations of Lamarckism Vienna Series in Theoretical Biology Gerd B
Transformations of Lamarckism Vienna Series in Theoretical Biology Gerd B. M ü ller, G ü nter P. Wagner, and Werner Callebaut, editors The Evolution of Cognition , edited by Cecilia Heyes and Ludwig Huber, 2000 Origination of Organismal Form: Beyond the Gene in Development and Evolutionary Biology , edited by Gerd B. M ü ller and Stuart A. Newman, 2003 Environment, Development, and Evolution: Toward a Synthesis , edited by Brian K. Hall, Roy D. Pearson, and Gerd B. M ü ller, 2004 Evolution of Communication Systems: A Comparative Approach , edited by D. Kimbrough Oller and Ulrike Griebel, 2004 Modularity: Understanding the Development and Evolution of Natural Complex Systems , edited by Werner Callebaut and Diego Rasskin-Gutman, 2005 Compositional Evolution: The Impact of Sex, Symbiosis, and Modularity on the Gradualist Framework of Evolution , by Richard A. Watson, 2006 Biological Emergences: Evolution by Natural Experiment , by Robert G. B. Reid, 2007 Modeling Biology: Structure, Behaviors, Evolution , edited by Manfred D. Laubichler and Gerd B. M ü ller, 2007 Evolution of Communicative Flexibility: Complexity, Creativity, and Adaptability in Human and Animal Communication , edited by Kimbrough D. Oller and Ulrike Griebel, 2008 Functions in Biological and Artifi cial Worlds: Comparative Philosophical Perspectives , edited by Ulrich Krohs and Peter Kroes, 2009 Cognitive Biology: Evolutionary and Developmental Perspectives on Mind, Brain, and Behavior , edited by Luca Tommasi, Mary A. Peterson, and Lynn Nadel, 2009 Innovation in Cultural Systems: Contributions from Evolutionary Anthropology , edited by Michael J. O ’ Brien and Stephen J. Shennan, 2010 The Major Transitions in Evolution Revisited , edited by Brett Calcott and Kim Sterelny, 2011 Transformations of Lamarckism: From Subtle Fluids to Molecular Biology , edited by Snait B. -

Introduction to the Cell Cell History Cell Structures and Functions
Introduction to the cell cell history cell structures and functions CK-12 Foundation December 16, 2009 CK-12 Foundation is a non-profit organization with a mission to reduce the cost of textbook materials for the K-12 market both in the U.S. and worldwide. Using an open-content, web-based collaborative model termed the “FlexBook,” CK-12 intends to pioneer the generation and distribution of high quality educational content that will serve both as core text as well as provide an adaptive environment for learning. Copyright ©2009 CK-12 Foundation This work is licensed under the Creative Commons Attribution-Share Alike 3.0 United States License. To view a copy of this license, visit http://creativecommons.org/licenses/by-sa/3.0/us/ or send a letter to Creative Commons, 171 Second Street, Suite 300, San Francisco, California, 94105, USA. Contents 1 Cell structure and function dec 16 5 1.1 Lesson 3.1: Introduction to Cells .................................. 5 3 www.ck12.org www.ck12.org 4 Chapter 1 Cell structure and function dec 16 1.1 Lesson 3.1: Introduction to Cells Lesson Objectives • Identify the scientists that first observed cells. • Outline the importance of microscopes in the discovery of cells. • Summarize what the cell theory proposes. • Identify the limitations on cell size. • Identify the four parts common to all cells. • Compare prokaryotic and eukaryotic cells. Introduction Knowing the make up of cells and how cells work is necessary to all of the biological sciences. Learning about the similarities and differences between cell types is particularly important to the fields of cell biology and molecular biology. -

The Act of Controlling Adult Stem Cell Dynamics: Insights from Animal Models
biomolecules Review The Act of Controlling Adult Stem Cell Dynamics: Insights from Animal Models Meera Krishnan 1, Sahil Kumar 1, Luis Johnson Kangale 2,3 , Eric Ghigo 3,4 and Prasad Abnave 1,* 1 Regional Centre for Biotechnology, NCR Biotech Science Cluster, 3rd Milestone, Gurgaon-Faridabad Ex-pressway, Faridabad 121001, India; [email protected] (M.K.); [email protected] (S.K.) 2 IRD, AP-HM, SSA, VITROME, Aix-Marseille University, 13385 Marseille, France; [email protected] 3 Institut Hospitalo Universitaire Méditerranée Infection, 13385 Marseille, France; [email protected] 4 TechnoJouvence, 13385 Marseille, France * Correspondence: [email protected] Abstract: Adult stem cells (ASCs) are the undifferentiated cells that possess self-renewal and differ- entiation abilities. They are present in all major organ systems of the body and are uniquely reserved there during development for tissue maintenance during homeostasis, injury, and infection. They do so by promptly modulating the dynamics of proliferation, differentiation, survival, and migration. Any imbalance in these processes may result in regeneration failure or developing cancer. Hence, the dynamics of these various behaviors of ASCs need to always be precisely controlled. Several genetic and epigenetic factors have been demonstrated to be involved in tightly regulating the proliferation, differentiation, and self-renewal of ASCs. Understanding these mechanisms is of great importance, given the role of stem cells in regenerative medicine. Investigations on various animal models have played a significant part in enriching our knowledge and giving In Vivo in-sight into such ASCs regulatory mechanisms. In this review, we have discussed the recent In Vivo studies demonstrating the role of various genetic factors in regulating dynamics of different ASCs viz. -

Epigenetic Metabolites License Stem Cell States
CHAPTER SIX Epigenetic metabolites license stem cell states Logeshwaran Somasundarama,b,†, Shiri Levya,b,†, Abdiasis M. Husseina,b, Devon D. Ehnesa,b, Julie Mathieub,c, Hannele Ruohola-Bakera,b,∗ aDepartment of Biochemistry, University of Washington, Seattle, WA, United States bInstitute for Stem Cell and Regenerative Medicine, University of Washington, Seattle, WA, United States cDepartment of Comparative Medicine, University of Washington, Seattle, WA, United States ∗ Corresponding author: e-mail address: [email protected] Contents 1. Introduction 210 2. Stem cell energetics 210 3. Metabolism of quiescent stem cells 212 3.1 Adult stem cells 212 3.2 Pluripotent stem cell quiescence, diapause 216 4. Metabolism of active stem cells 217 4.1 Metabolism after fertilization 217 4.2 Metabolism of pre-implantation and post-implantation pluripotent stem cells 218 4.3 Metabolism of actively cycling adult stem cells: MSC as case-study 220 5. HIF, the master regulator of metabolism 222 6. Epigenetic signatures and epigenetic metabolites 224 6.1 Epigenetic signatures of naïve and primed pluripotent stem cells 224 6.2 Epigenetic signatures of adult stem cells 227 6.3 Epigenetic metabolites 228 7. Conclusion 229 Acknowledgments 230 References 230 Further reading 240 Abstract It has become clear during recent years that stem cells undergo metabolic remodeling during their activation process. While these metabolic switches take place in pluripotency as well as adult stem cell populations, the rules that govern the switch are not clear. † Equal contribution. # Current Topics in Developmental Biology, Volume 138 2020 Elsevier Inc. 209 ISSN 0070-2153 All rights reserved. https://doi.org/10.1016/bs.ctdb.2020.02.003 210 Logeshwaran Somasundaram et al. -

DNA Methylation, Imprinting and Cancer
European Journal of Human Genetics (2002) 10, 6±16 ã 2002 Nature Publishing Group All rights reserved 1018-4813/02 $25.00 www.nature.com/ejhg REVIEW DNA methylation, imprinting and cancer Christoph Plass*,1 and Paul D Soloway*,2 1Division of Human Cancer Genetics and the Comprehensive Cancer Center, The Ohio State University, Columbus, Ohio, OH 43210, USA; 2Department of Molecular and Cellular Biology, Roswell Park Cancer Institute, Buffalo, New York, NY 14263, USA It is well known that a variety of genetic changes influence the development and progression of cancer. These changes may result from inherited or spontaneous mutations that are not corrected by repair mechanisms prior to DNA replication. It is increasingly clear that so called epigenetic effects that do not affect the primary sequence of the genome also play an important role in tumorigenesis. This was supported initially by observations that cancer genomes undergo changes in their methylation state and that control of parental allele-specific methylation and expression of imprinted loci is lost in several cancers. Many loci acquiring aberrant methylation in cancers have since been identified and shown to be silenced by DNA methylation. In many cases, this mechanism of silencing inactivates tumour suppressors as effectively as frank mutation and is one of the cancer-predisposing hits described in Knudson's two hit hypothesis. In contrast to mutations which are essentially irreversible, methylation changes are reversible, raising the possibility of developing therapeutics based on restoring the normal methylation state to cancer-associated genes. Development of such therapeutics will require identifying loci undergoing methylation changes in cancer, understanding how their methylation influences tumorigenesis and identifying the mechanisms regulating the methylation state of the genome. -
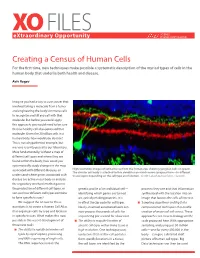
Creating a Census of Human Cells
XO FILES SCIENCE eXtraordinary Opportunity PHILANTHROPY ALLIANCE Creating a Census of Human Cells For the first time, new techniques make possible a systematic description of the myriad types of cells in the human body that underlie both health and disease. Aviv Regev Imagine you had a way to cure cancer that involved taking a molecule from a tumor and engineering the body’s immune cells to recognize and kill any cell with that molecule. But before you could apply this approach, you would need to be sure that no healthy cell also expressed that molecule. Given the 20 trillion cells in a human body, how would you do that? This is not a hypothetical example, but was one recently posed to our laboratory. More fundamentally, without a map of different cell types and where they are found within the body, how could you systematically study changes in the map associated with different diseases, or High resolution images of retinal tissue from the human eye, showing ganglion cells (in green). The circular cell body is attached to thin dendrites on which nerve synapses form—in different understand where genes associated with tissue layers depending on the cell type and function. Credit: Lab of Joshua Sanes, Harvard. disease are active in our body or analyze the regulatory mechanisms that govern the production of different cell types, or genetic profile of an individual cell— proteins they use and that information sort out how different cell types combine identifying which genes are turned synthesized with the location into an to form specific tissues? on, actively making proteins. -

Robustness and Timing of Cellular Differentiation Through Population-Based Symmetry Breaking
AR TI CLE Robustness AND TIMING OF CELLULAR DIFFERENTIATION THROUGH population-based SYMMETRY BREAKING Angel StanoeV1 | Christian Schröter1 | Aneta KOSESKA1∗ 1Department OF Systemic Cell Biology, Max Planck Institute OF Molecular PhYSIOLOGY, Although COORDINATED BEHAVIOR OF MULTICELLULAR ORGANISMS Dortmund, GermanY RELIES ON INTERCELLULAR communication, THE DYNAMICAL mech- Correspondence ANISM THAT UNDERLIES SYMMETRY BREAKING IN HOMOGENEOUS Email: populations, THEREBY GIVING RISE TO HETEROGENEOUS CELL TYPES ∗[email protected] REMAINS UNCLEAR. In CONTRAST TO THE PREVALENT cell-intrinsic VIEW OF DIFFERENTIATION WHERE MULTISTABILITY ON THE SINGLE CELL LEVEL IS A NECESSARY pre-requisite, WE PROPOSE THAT het- EROGENEOUS CELLULAR IDENTITIES EMERGE AND ARE MAINTAINED COOPERATIVELY ON A POPULATION LEVel. Identifying A GENERIC SYMMETRY BREAKING mechanism, WE DEMONSTRATE THAT ROBUST PROPORTIONS BETWEEN DIFFERENTIATED CELL TYPES ARE INTRINSIC FEATURES OF THIS INHOMOGENEOUS STEADY STATE solution. Crit- ICAL ORGANIZATION IN THE VICINITY OF THE SYMMETRY BREAKING BIFURCATION ON THE OTHER hand, SUGGESTS THAT TIMING OF cel- LULAR DIFFERENTIATION EMERGES FOR GROWING POPULATIONS IN A self-organized MANNER. Setting A CLEAR DISTINCTION BETWEEN pre-eXISTING ASYMMETRIES AND SYMMETRY BREAKING EVents, WE THUS DEMONSTRATE THAT EMERGENT SYMMETRY breaking, IN CONJUNCTION WITH ORGANIZATION NEAR CRITICALITY NECESSARILY LEADS TO ROBUSTNESS AND ACCURACY DURING DEVelopment. KEYWORDS SYMMETRY breaking; differentiation; INHOMOGENEOUS STEADY STATE Abbreviations: -

Translational Applications of Adult Stem Cell-Derived Organoids Jarno Drost1,2,* and Hans Clevers1,2,3,‡
© 2017. Published by The Company of Biologists Ltd | Development (2017) 144, 968-975 doi:10.1242/dev.140566 PRIMER Translational applications of adult stem cell-derived organoids Jarno Drost1,2,* and Hans Clevers1,2,3,‡ ABSTRACT of organotypic organoids from single Lgr5-positive ISCs (Sato Adult stem cells from a variety of organs can be expanded long- et al., 2009). These organoids contained all cell types of the ‘ ’ term in vitro as three-dimensional organotypic structures termed intestinal epithelium and were structured into proliferative crypt ‘ ’ organoids. These adult stem cell-derived organoids retain their organ and differentiated villus compartments, thereby retaining the identity and remain genetically stable over long periods of time. The architecture of the native intestine (Sato et al., 2009). In addition ability to grow organoids from patient-derived healthy and diseased to Wnt/R-spondin, epidermal growth factor (EGF) (Dignass and tissue allows for the study of organ development, tissue homeostasis Sturm, 2001), noggin (BMP inhibitor) (Haramis et al., 2004) and an and disease. In this Review, we discuss the generation of adult artificial laminin-rich extracellular matrix (provided by Matrigel) stem cell-derived organoid cultures and their applications in in vitro complemented the cocktail for successful in vitro propagation of disease modeling, personalized cancer therapy and regenerative mouse ISC-derived organoids (Sato et al., 2009). At the same time, medicine. Ootani et al. reported another Wnt-driven in vitro culture of ISCs (Ootani et al., 2009). In contrast to Sato et al., this culture was not KEY WORDS: Adult stem cells, Organoids, Disease modelling, based on defined growth factors.