Cell-Type-Specific Expression Quantitative Trait Loci Associated with Alzheimer Disease in Blood and Brain Tissue
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Introduction to the Cell Cell History Cell Structures and Functions
Introduction to the cell cell history cell structures and functions CK-12 Foundation December 16, 2009 CK-12 Foundation is a non-profit organization with a mission to reduce the cost of textbook materials for the K-12 market both in the U.S. and worldwide. Using an open-content, web-based collaborative model termed the “FlexBook,” CK-12 intends to pioneer the generation and distribution of high quality educational content that will serve both as core text as well as provide an adaptive environment for learning. Copyright ©2009 CK-12 Foundation This work is licensed under the Creative Commons Attribution-Share Alike 3.0 United States License. To view a copy of this license, visit http://creativecommons.org/licenses/by-sa/3.0/us/ or send a letter to Creative Commons, 171 Second Street, Suite 300, San Francisco, California, 94105, USA. Contents 1 Cell structure and function dec 16 5 1.1 Lesson 3.1: Introduction to Cells .................................. 5 3 www.ck12.org www.ck12.org 4 Chapter 1 Cell structure and function dec 16 1.1 Lesson 3.1: Introduction to Cells Lesson Objectives • Identify the scientists that first observed cells. • Outline the importance of microscopes in the discovery of cells. • Summarize what the cell theory proposes. • Identify the limitations on cell size. • Identify the four parts common to all cells. • Compare prokaryotic and eukaryotic cells. Introduction Knowing the make up of cells and how cells work is necessary to all of the biological sciences. Learning about the similarities and differences between cell types is particularly important to the fields of cell biology and molecular biology. -

PILRA Antibody Cat
PILRA Antibody Cat. No.: 30-252 PILRA Antibody Specifications HOST SPECIES: Rabbit SPECIES REACTIVITY: Human Antibody produced in rabbits immunized with a synthetic peptide corresponding a region IMMUNOGEN: of human PILRA. TESTED APPLICATIONS: ELISA, WB PILRA antibody can be used for detection of PILRA by ELISA at 1:1562500. PILRA antibody APPLICATIONS: can be used for detection of PILRA by western blot at 0.5 μg/mL, and HRP conjugated secondary antibody should be diluted 1:50,000 - 100,000. POSITIVE CONTROL: 1) Cat. No. 1211 - HepG2 Cell Lysate PREDICTED MOLECULAR 18 kDa WEIGHT: Properties PURIFICATION: Antibody is purified by peptide affinity chromatography method. CLONALITY: Polyclonal CONJUGATE: Unconjugated PHYSICAL STATE: Liquid September 25, 2021 1 https://www.prosci-inc.com/pilra-antibody-30-252.html Purified antibody supplied in 1x PBS buffer with 0.09% (w/v) sodium azide and 2% BUFFER: sucrose. CONCENTRATION: batch dependent For short periods of storage (days) store at 4˚C. For longer periods of storage, store PILRA STORAGE CONDITIONS: antibody at -20˚C. As with any antibody avoid repeat freeze-thaw cycles. Additional Info OFFICIAL SYMBOL: PILRA ALTERNATE NAMES: PILRA, FDF03 ACCESSION NO.: NP_840057 PROTEIN GI NO.: 30179907 GENE ID: 29992 USER NOTE: Optimal dilutions for each application to be determined by the researcher. Background and References Cell signaling pathways rely on a dynamic interaction between activating and inhibiting processes. SHP-1-mediated dephosphorylation of protein tyrosine residues is central to the regulation of several cell signaling pathways. Two types of inhibitory receptor superfamily members are immunoreceptor tyrosine-based inhibitory motif (ITIM)-bearing receptors and their non-ITIM-bearing, activating counterparts. -
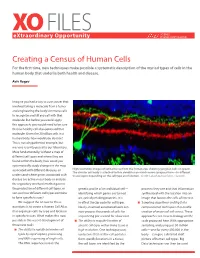
Creating a Census of Human Cells
XO FILES SCIENCE eXtraordinary Opportunity PHILANTHROPY ALLIANCE Creating a Census of Human Cells For the first time, new techniques make possible a systematic description of the myriad types of cells in the human body that underlie both health and disease. Aviv Regev Imagine you had a way to cure cancer that involved taking a molecule from a tumor and engineering the body’s immune cells to recognize and kill any cell with that molecule. But before you could apply this approach, you would need to be sure that no healthy cell also expressed that molecule. Given the 20 trillion cells in a human body, how would you do that? This is not a hypothetical example, but was one recently posed to our laboratory. More fundamentally, without a map of different cell types and where they are found within the body, how could you systematically study changes in the map associated with different diseases, or High resolution images of retinal tissue from the human eye, showing ganglion cells (in green). The circular cell body is attached to thin dendrites on which nerve synapses form—in different understand where genes associated with tissue layers depending on the cell type and function. Credit: Lab of Joshua Sanes, Harvard. disease are active in our body or analyze the regulatory mechanisms that govern the production of different cell types, or genetic profile of an individual cell— proteins they use and that information sort out how different cell types combine identifying which genes are turned synthesized with the location into an to form specific tissues? on, actively making proteins. -

Robustness and Timing of Cellular Differentiation Through Population-Based Symmetry Breaking
AR TI CLE Robustness AND TIMING OF CELLULAR DIFFERENTIATION THROUGH population-based SYMMETRY BREAKING Angel StanoeV1 | Christian Schröter1 | Aneta KOSESKA1∗ 1Department OF Systemic Cell Biology, Max Planck Institute OF Molecular PhYSIOLOGY, Although COORDINATED BEHAVIOR OF MULTICELLULAR ORGANISMS Dortmund, GermanY RELIES ON INTERCELLULAR communication, THE DYNAMICAL mech- Correspondence ANISM THAT UNDERLIES SYMMETRY BREAKING IN HOMOGENEOUS Email: populations, THEREBY GIVING RISE TO HETEROGENEOUS CELL TYPES ∗[email protected] REMAINS UNCLEAR. In CONTRAST TO THE PREVALENT cell-intrinsic VIEW OF DIFFERENTIATION WHERE MULTISTABILITY ON THE SINGLE CELL LEVEL IS A NECESSARY pre-requisite, WE PROPOSE THAT het- EROGENEOUS CELLULAR IDENTITIES EMERGE AND ARE MAINTAINED COOPERATIVELY ON A POPULATION LEVel. Identifying A GENERIC SYMMETRY BREAKING mechanism, WE DEMONSTRATE THAT ROBUST PROPORTIONS BETWEEN DIFFERENTIATED CELL TYPES ARE INTRINSIC FEATURES OF THIS INHOMOGENEOUS STEADY STATE solution. Crit- ICAL ORGANIZATION IN THE VICINITY OF THE SYMMETRY BREAKING BIFURCATION ON THE OTHER hand, SUGGESTS THAT TIMING OF cel- LULAR DIFFERENTIATION EMERGES FOR GROWING POPULATIONS IN A self-organized MANNER. Setting A CLEAR DISTINCTION BETWEEN pre-eXISTING ASYMMETRIES AND SYMMETRY BREAKING EVents, WE THUS DEMONSTRATE THAT EMERGENT SYMMETRY breaking, IN CONJUNCTION WITH ORGANIZATION NEAR CRITICALITY NECESSARILY LEADS TO ROBUSTNESS AND ACCURACY DURING DEVelopment. KEYWORDS SYMMETRY breaking; differentiation; INHOMOGENEOUS STEADY STATE Abbreviations: -

Blood Flow Guides Sequential Support of Neutrophil Arrest and Diapedesis
RESEARCH ARTICLE Blood flow guides sequential support of neutrophil arrest and diapedesis by PILR-b 1 and PILR-a Yu-Tung Li, Debashree Goswami†, Melissa Follmer, Annette Artz, Mariana Pacheco-Blanco, Dietmar Vestweber* Vascular Cell Biology, Max Planck Institute of Molecular Biomedicine, Mu¨ nster, Germany Abstract Arrest of rapidly flowing neutrophils in venules relies on capturing through selectins and chemokine-induced integrin activation. Despite a long-established concept, we show here that gene inactivation of activating paired immunoglobulin-like receptor (PILR)-b1 nearly halved the efficiency of neutrophil arrest in venules of the mouse cremaster muscle. We found that this receptor binds to CD99, an interaction which relies on flow-induced shear forces and boosts chemokine-induced b2-integrin-activation, leading to neutrophil attachment to endothelium. Upon arrest, binding of PILR-b1 to CD99 ceases, shifting the signaling balance towards inhibitory PILR-a. This enables integrin deactivation and supports cell migration. Thus, flow-driven shear forces guide sequential signaling of first activating PILR-b1 followed by inhibitory PILR-a to prompt neutrophil arrest and then transmigration. This doubles the efficiency of selectin-chemokine driven neutrophil arrest by PILR-b1 and then supports transition to migration by PILR-a. DOI: https://doi.org/10.7554/eLife.47642.001 *For correspondence: [email protected] Present address: †Center for Global Infectious Disease Introduction Research, Seattle Childrens Host defense against pathogens depends on the recruitment of leukocytes to sites of infections Research Institute, Seattle, (Ley et al., 2007). Selectins capture leukocytes to the endothelial cell surface by binding to glyco- United States conjugates (McEver, 2015). -

A Concise Review on the Classification and Nomenclature of Stem Cells Kök Hücrelerinin S›N›Fland›R›Lmas› Ve Isimlendirilmesine Iliflkin K›Sa Bir Derleme
Review 57 A concise review on the classification and nomenclature of stem cells Kök hücrelerinin s›n›fland›r›lmas› ve isimlendirilmesine iliflkin k›sa bir derleme Alp Can Ankara University Medical School, Department of Histology and Embryology, Ankara, Turkey Abstract Stem cell biology and regenerative medicine is a relatively young field. However, in recent years there has been a tremen- dous interest in stem cells possibly due to their therapeutic potential in disease states. As a classical definition, a stem cell is an undifferentiated cell that can produce daughter cells that can either remain a stem cell in a process called self-renew- al, or commit to a specific cell type via the initiation of a differentiation pathway leading to the production of mature progeny cells. Despite this acknowledged definition, the classification of stem cells has been a perplexing notion that may often raise misconception even among stem cell biologists. Therefore, the aim of this brief review is to give a conceptual approach to classifying the stem cells beginning from the early morula stage totipotent embryonic stem cells to the unipotent tissue-resident adult stem cells, also called tissue-specific stem cells. (Turk J Hematol 2008; 25: 57-9) Key words: Stem cells, embryonic stem cells, tissue-specific stem cells, classification, progeny. Özet Kök hücresi biyolojisi ve onar›msal t›p görece yeni alanlard›r. Buna karfl›n, son y›llarda çeflitli hastal›klarda tedavi amac›yla kullan›labilme potansiyelleri nedeniyle kök hücrelerine ola¤anüstü bir ilgi art›fl› vard›r. Klasik tan›m›yla kök hücresi, kendini yenileme ad› verilen mekanizmayla farkl›laflmadan kendini ço¤altan veya bir dizi farkl›laflma aflamas›ndan geçerek olgun hücrelere dönüflebilen hücrelerdir. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Myosin 1E Interacts with Synaptojanin-1 and Dynamin and Is Involved in Endocytosis
FEBS Letters 581 (2007) 644–650 Myosin 1E interacts with synaptojanin-1 and dynamin and is involved in endocytosis Mira Krendela,*, Emily K. Osterweila, Mark S. Moosekera,b,c a Department of Molecular, Cellular, and Developmental Biology, Yale University, New Haven, CT 06511, USA b Department of Cell Biology, Yale University, New Haven, CT 06511, USA c Department of Pathology, Yale University, New Haven, CT 06511, USA Received 21 November 2006; revised 8 January 2007; accepted 11 January 2007 Available online 18 January 2007 Edited by Felix Wieland Myo1 isoforms (Myo3p and Myo5p) leads to defects in endo- Abstract Myosin 1E is one of two ‘‘long-tailed’’ human Class I myosins that contain an SH3 domain within the tail region. SH3 cytosis [3].InAcanthamoeba, various Myo1 isoforms are domains of yeast and amoeboid myosins I interact with activa- found in association with intracellular vesicles [10].InDictyos- tors of the Arp2/3 complex, an important regulator of actin poly- telium, long-tailed Myo1s (myo B, C, and D) are required for merization. No binding partners for the SH3 domains of myosins fluid-phase endocytosis [11]. I have been identified in higher eukaryotes. In the current study, Myo1e, the mouse homolog of the human long-tailed myo- we show that two proteins with prominent functions in endocyto- sin, Myo1E (formerly referred to as Myo1C under the old myo- sis, synaptojanin-1 and dynamin, bind to the SH3 domain of sin nomenclature [12]), has been previously localized to human Myo1E. Myosin 1E co-localizes with clathrin- and dyn- phagocytic structures [13]. In this study, we report that Myo1E amin-containing puncta at the plasma membrane and this co- binds to two proline-rich proteins, synaptojanin-1 and dyn- localization requires an intact SH3 domain. -

Epistatic Interactions of Genetic Loci Associated with Age-Related
www.nature.com/scientificreports OPEN Epistatic interactions of genetic loci associated with age‑related macular degeneration Christina Kiel1,3, Christoph A. Nebauer1,3, Tobias Strunz1,3, Simon Stelzl1 & Bernhard H. F. Weber 1,2* The currently largest genome‑wide association study (GWAS) for age‑related macular degeneration (AMD) defnes disease association with genome‑wide signifcance for 52 independent common and rare genetic variants across 34 chromosomal loci. Overall, these loci contain over 7200 variants and are enriched for genes with functions indicating several shared cellular processes. Still, the precise mechanisms leading to AMD pathology are largely unknown. Here, we exploit the phenomenon of epistatic interaction to identify seemingly independent AMD‑associated variants that reveal joint efects on gene expression. We focus on genetic variants associated with lipid metabolism, organization of extracellular structures, and innate immunity, specifcally the complement cascade. Multiple combinations of independent variants were used to generate genetic risk scores allowing gene expression in liver to be compared between low and high‑risk AMD. We identifed genetic variant combinations correlating signifcantly with expression of 26 genes, of which 19 have not been associated with AMD before. This study defnes novel targets and allows prioritizing further functional work into AMD pathobiology. A frst successful genome-wide association study (GWAS) was reported in 2005 and identifed with genome- wide signifcance genetic variants at the CFH locus associated with age-related macular degeneration (AMD), a complex disease which is a frequent cause of progressive vision loss in the elderly population 1. Since then, the list of AMD-associated genetic variation has grown exponentially, presently bringing the total to 52 independent common and rare variants across 34 chromosomal loci2. -

Genome-Wide Identification, Characterization and Expression
G C A T T A C G G C A T genes Article Genome-Wide Identification, Characterization and Expression Profiling of myosin Family Genes in Sebastes schlegelii Chaofan Jin 1, Mengya Wang 1,2, Weihao Song 1 , Xiangfu Kong 1, Fengyan Zhang 1, Quanqi Zhang 1,2,3 and Yan He 1,2,* 1 MOE Key Laboratory of Molecular Genetics and Breeding, College of Marine Life Sciences, Ocean University of China, Qingdao 266003, China; [email protected] (C.J.); [email protected] (M.W.); [email protected] (W.S.); [email protected] (X.K.); [email protected] (F.Z.); [email protected] (Q.Z.) 2 Laboratory of Tropical Marine Germplasm Resources and Breeding Engineering, Sanya Oceanographic Institution, Ocean University of China, Sanya 572000, China 3 Laboratory for Marine Fisheries Science and Food Production Processes, Qingdao National Laboratory for Marine Science and Technology, Qingdao 266003, China * Correspondence: [email protected]; Tel.: +86-0532-82031986 Abstract: Myosins are important eukaryotic motor proteins that bind actin and utilize the energy of ATP hydrolysis to perform a broad range of functions such as muscle contraction, cell migration, cytokinesis, and intracellular trafficking. However, the characterization and function of myosin is poorly studied in teleost fish. In this study, we identified 60 myosin family genes in a marine teleost, black rockfish (Sebastes schlegelii), and further characterized their expression patterns. myosin showed divergent expression patterns in adult tissues, indicating they are involved in different types and Citation: Jin, C.; Wang, M.; Song, W.; compositions of muscle fibers. Among 12 subfamilies, S. schlegelii myo2 subfamily was significantly Kong, X.; Zhang, F.; Zhang, Q.; He, Y. -

Genetic Variability in a Holstein Population Using SNP Markers and Their Use for Monitoring Mating Strategies
https://doi.org/10.22319/rmcp.v10i3.4842 Article Genetic variability in a Holstein population using SNP markers and their use for monitoring mating strategies Kathy Scienski a,b,c Angelo Ialacci c Alessandro Bagnato c Davide Reginelli d Marina Durán-Aguilar e Maria Giuseppina Strillacci c* a Texas A&M University, College Station. Interdisciplinary Program in Genetics. Texas, USA. b Texas A&M University. Department of Animal Science, Texas, USA. c Università degli Studi di Milano. Department of Veterinary Medicine, Via Trentacoste 2, 20134 Milano, Italy. d Università degli Studi di Milano. Azienda Agraria Didattico Sperimentale Angelo Menozzi, Landriano, Pavia, Italy. e Universidad Autónoma de Querétaro. Facultad de Ciencias Naturales. Querétaro. México. * Corresponding author: [email protected] Abstract: As genotyping costs continue to decrease, the demand for genotyping has increased among farmers. In most livestock herds, an important issue is controlling the increase in inbreeding coefficient. While this remains a large motive to genotype, producers are often unaware of the other benefits that genotyping could bring. The aim of this study was to demonstrate that SNP chips could be used as an effective herd management tool by utilizing a population of Italian Holstein-Friesian cattle. After filtering, the total number 643 Rev Mex Cienc Pecu 2019;10(3):643-663 of animals and SNPs retained for analyses were 44 and 27,365, respectively. The principal component analyses (PCA) were able to identify a sire and origin-of-sire effect within the herd, while determining that sires do not influence individual genomic selection index values. The inbreeding coefficients calculated from genotypes (FIS) provided a glimpse into the herd’s heterozygosity and determined that the genetic variability is being well maintained.