Cell Wall Biology in Oomycete Pathogenicity
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Assessment of Forest Pests and Diseases in Protected Areas of Georgia Final Report
Assessment of Forest Pests and Diseases in Protected Areas of Georgia Final report Dr. Iryna Matsiakh Tbilisi 2014 This publication has been produced with the assistance of the European Union. The content, findings, interpretations, and conclusions of this publication are the sole responsibility of the FLEG II (ENPI East) Programme Team (www.enpi-fleg.org) and can in no way be taken to reflect the views of the European Union. The views expressed do not necessarily reflect those of the Implementing Organizations. CONTENTS LIST OF TABLES AND FIGURES ............................................................................................................................. 3 ABBREVIATIONS AND ACRONYMS ...................................................................................................................... 6 EXECUTIVE SUMMARY .............................................................................................................................................. 7 Background information ...................................................................................................................................... 7 Literature review ...................................................................................................................................................... 7 Methodology ................................................................................................................................................................. 8 Results and Discussion .......................................................................................................................................... -
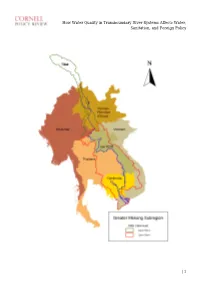
How Water Quality in Transboundary River Systems Affects Water, Sanitation, and Foreign Policy
How Water Quality in Transboundary River Systems Affects Water, Sanitation, and Foreign Policy | 1 How Water Quality in Transboundary River Systems Affects Water, Sanitation, and Foreign Policy David Tipping, 2001 By David C. Tipping Edited by Yeareen Yun Disclaimer: The views and opinions expressed in this article are those of the author and do not necessarily reflect the official policy or position of any agency of the Australian government. Assumptions made within the analysis are not reflective of the position of any Australian government entity, or other organization or professional association. 1. INTRODUCTION Access to adequate water supply and sanitation is the core premise of local level water security. Effective management of transboundary river basin systems and water quality risks is therefore fundamental to social progress and quality of life. Improved water quality management benefits many individual lives in riparian nations, and, as demonstrated by the annual new year blessing of the fish migrations, society at large throughout the Mekong River Basin. In 2001, the author investigated the use of sustainable development indicators to improve the institutional effectiveness of international environmental management regimes. A new framework was designed to evaluate beneficial uses of water. In addition, a case study was developed on the Lower Mekong River Basin system, which integrated measures of water and environmental quality and socio-economic development. The research objectives were: (1) improving the understanding of water quality issues; (2) benchmarking water resources management performance at local, national and regional levels; and (3) enhancing technical and administrative capabilities of transboundary river basin management regimes through capacity development focused on the achievement of sustainable development objectives, and obligations and duties under international law. -

Assessment of Forest Pests and Diseases in Native Boxwood Forests of Georgia Final Report
Assessment of Forest Pests and Diseases in Native Boxwood Forests of Georgia Final report Dr. Iryna Matsiakh Forestry Department, Ukrainian National Forestry University (Lviv) Tbilisi 2016 TABLE OF CONTENT LIST OF TABLES AND FIGURES .................................................................................................................................. 2 ABBREVIATIONS AND ACRONYMS ........................................................................................................................... 5 EXECUTIVE SUMMARY .................................................................................................................................................. 6 INTRODUCTION .............................................................................................................................................................. 10 1. BACKGROUND INFORMATION ............................................................................................................................ 11 1.1. Biodiversity of Georgia ........................................................................................................................................ 11 1.2. Forest Ecosystems .................................................................................................................................................. 12 1.3. Boxwood Forests in Forests Habitat Classification ................................................................................. 14 1.4. Georgian Forests Habitat in the Context of Climate Change -

Guide 3 – Fish Farmer's Guide to Combating Parasitic
GUIDE 3 – FISH FARMER’S GUIDE TO COMBATING PARASITIC INFECTIONS IN COMMON CARP AQUACULTURE e-NIPO: 833-20-103-X A Series of ParaFishControl Guides to Combating Fish Parasite Infections in Aquaculture. Guide 3 This project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No. 634429 (ParaFishControl). This output reflects only the author’s view and the European Union cannot be held responsible for any use that may be made of the information contained therein. Wherever the fish are, that's where we go. “ Richard Wagner “ Common carp is the third most cultivated freshwater species in the world. Carp aquaculture is usually performed in a semi-intensive manner, in earthen ponds, where parasitic diseases can easily compromise fish health, especially in the hot summer months, leading to production and economic losses. This guide provides useful information about the biological background of five parasites, their diagnostics and control measures. © A.S. Holzer List of Authors Dr Astrid S. Holzer, Principal Investigator and Team Leader Institute of Parasitology Biology Centre of the Czech Academy of Sciences, Czech Republic Email: [email protected] Dr Pavla Bartošová-Sojková, Researcher Institute of Parasitology Biology Centre of the Czech Academy of Sciences, Czech Republic Email: [email protected] Honorary Prof. Csaba Székely, Scientific Advisor and Team Leader Institute for Veterinary Medical Research, Centre for Agricultural Research, (former Hungarian Academy of Sciences), Hungary Email: [email protected] Dr Gábor Cech, Senior Researcher, Institute for Veterinary Medical Research, Centre for Agricultural Research, (former Hungarian Academy of Sciences), Hungary Email: [email protected] Dr Kálmán Molnár, Retired Scientific Advisor, Fish Pathology and Parasitology Research Team, Institute for Veterinary Medical Research, Centre for Agricultural Research (former Hungarian Academy of Sciences), Hungary Prof. -

Difference Between Haplontic and Diplontic Life Cycles
Difference Between Haplontic and Diplontic Life Cycles www.differencebetween.com Key Difference – Haplontic vs Diplontic Life Cycles In the context of biology, a biological life cycle is a sequence of changes a particular organism undergoes through means of reproduction (sexual or asexual) which finally returns to the original starting phase. This procedure differs from one organism to the other. During sexual reproduction, the life cycle includes the change of ploidy; the alternation of haploid (n) and diploid (2n) stages. Meiosis occurs during the change over from a diploid stage to a haploid stage. With regards to change of ploidy, life cycles are of three types. They are, haplontic, diplontic and haplodiplontic. In a haplontic life cycle, the haploid stage is typically multicellular and results in the formation of a diploid (2n) cell, which is a zygote. The zygote undergoes meiosis, which results in the formation of haploid (n) cells. In a diplontic life cycle, the diploid stage is typically multicellular, and meiosis occurs during gamete formation which results in the production of haploid (n) gametes. During fertilization, the haploid (n) gametes fuse together in the formation of a diploid (2n) zygote, and it mitotically divides and produces a multicellular diploid (2n) organism. This is the key difference between haplontic and diplontic life cycles. What is a Haplontic Life Cycle? Haplontic life cycle involves the formation of a haploid (n) single cell by the meiosis of a diploid (2n) zygote. This phenomenon could be explained with sporic meiosis – the process of formation of spores. In this process, the zygote mitotically divides and produces multicellular sporophyte which is diploid (2n). -

FINAL COMPASS Aquaculture Roundtable Brief
Science in Action: Exploring the Future of U.S. Aquaculture A COMPASS Roundtable on Ocean Aquaculture As the population continues to expand—both domestically and globally—identifying secure, safe sources of protein is a critical need. With two-thirds of the planet covered in water, it is logical to turn to the ocean as an arena for producing food. Globally, aquaculture is the fastest growing food sector,[i] underscoring the importance of understanding the scientific, policy, and social implications of ocean aquaculture. As with all types of cultivated food production, there are complex and interwoven challenges and opportunities in ocean aquaculture.[ii] Indigenous knowledge and current research can answer questions around environmental safeguards, ecological impacts, long-term sustainable use of marine resources, and the social dimensions of ocean aquaculture. While we’ve developed a deeper scientific understanding of aquaculture, there remains a gap between the state of the science, federal policy, and public perceptions of ocean aquaculture in the U.S.[iii] In order to help provide research insights on the science related to aquaculture, COMPASS convened a roundtable discussion with scientists and policymakers in July 2019. The Roundtable examined ways that science can inform safe, sustainable, and socially acceptable ocean aquaculture in the United States. In preparation, COMPASS staff examined the U.S. aquaculture landscape by speaking with more than 50 scientists, managers, policymakers, and tribal representatives. These stage-setting conversations reflected the key concerns surrounding ocean aquaculture such as best management practices, economics, pollution, interactions with wild populations, and climate change. They also highlighted some of the scientific, technological, and cultural advancements in contemporary aquaculture that could address and reduce some of the perceived risks. -

Survivability of Moss and Fungal Spores in Tests Simulating
Biological Sciences in Space, Vol.25 No.2-4, 83-92,Takahashi, 2011 Y. et al. Original Paper Survivability of Moss and Fungal spores irradiated for 30 min. On the other hand, a colony did not develop if the spores were Spores in Tests Simulating taken off the beads and irradiated for 10 min. Conditions of the ISS Outer Wall This indicates that UV does not penetrate to the other side of the beads, and so the spores on Yuichi Takahashi1*, Hirofumi Hashimoto2, that side can be protected from UV radiation. 3 1 ©2011 Jpn. Soc. Biol. Sci. Space; Article ID: Takuo Nakagawa and Shinpei Shibata 112502011 1Department of Astrophysics, Interactive Symbio- sphere Science, Yamagata University Graduate Key words; fungi, moss, spore, survivability, thermal cycle treatment, UV irradiation School of Science and Engineering, Kojirakawa- machi 1-4-12, Yamagata-City, Yamagata 990-8560, Introduction Japan 2 Institute of Space and Astronautical Science, There are many factors that can influence the survivability JAXA, Yoshinodai 3-1-1 Chuo-ku, Sagamihara- of terrestrial life (TL) in interstellar and interplanetary space, City, Kanagawa 252-5210, Japan for example full-spectrum electromagnetic radiation from 3Inspection room, Kojirakawa-Shiseidoh Hospital, the sun, temperature fluctuations, cosmic particle radiation, Higashihara-machi 1-12-26, Yamagata-City, Ya- vacuum, and microgravity. However, space conditions can magata 990-0034, Japan only be partially simulated in ground experiments. Recently, several experiments have investigated the possibility of TL Abstract survival in space, for example EXPOSE-E (ESA Human Space Flight (2010) Expose, http:// smsc.cnes.fr/EXPOSE/), BIORISK (Baranow et al., 2009; Novikova et al., 2010) and To investigate whether terrestrial life (TL) can BIOPAN (De la Torre Noetzel et al., 2007; Devara et al., survive in interstellar and interplanetary space, 2010), while another one, TANPOPO, will take place in the an experiment was performed to simulate near future. -

Botany 12Th (Presented by Biology Wing DIET KUD H.O.D Sushma Gupta H.O.D Renuka Nagpal and Rashpal Singh Under the Guidance of Sh
1 | Page Botany 12th (Presented by Biology Wing DIET KUD H.O.D Sushma Gupta H.O.D Renuka Nagpal and Rashpal Singh Under the Guidance of Sh. Devinder Handoo H.O.D I/c DIET Kud) Unit-01 Reproduction in Flowering Plants Unit-01Marks- 07 ❖ Asexual reproduction Asexual reproduction is a mode of reproduction by which offspring arise from a single parent, and inherit the genes of that parent only, it is reproduction which does not involve meiosis, or fertilization. A more stringent definition is agamogenesis which is reproduction without the fusion of gametes. Asexual reproduction is the primary form of reproduction for single-celled organisms such as the archaea, bacteria, and protists. Many plants and fungi reproduce asexually as well. While all prokaryotes reproduce asexually (without the formation and fusion of gametes), mechanisms for lateral gene transfer such as conjugation, transformation and transduction are sometimes likened to sexual reproduction. A lack of sexual reproduction is relatively rare among multicellular organisms, particularly animals, for reasons that are not completely understood. Current hypotheses suggest that asexual reproduction may have short term benefits when rapid population growth is important or in stable environments, while sexual reproduction offers a net advantage by allowing more rapid generation of genetic diversity, allowing adaptation to changing environments. Developmental constraints may underlie why few animals have relinquished sexual reproduction completely in their life-cycles. Contents • 1 Types o 1.1 Binary fission o 1.2 Budding o 1.3 Vegetative reproduction o 1.4 Spore formation o 1.5 Fragmentation o 1.6 Parthenogenesis o 1.7 Agamogenesis o 1.8 Apomixis and nucellar embryony • 2 Alternation between sexual and asexual reproduction 2 | Page • 3 Examples in animals Types of Asexual Reproduction 1. -

The Biogeomorphological Life Cycle of Poplars During the Fluvial Biogeomorphological Succession: a Special Focus on Populus Nigra L
Open Archive TOULOUSE Archive Ouverte ( OATAO ) OATAO is an open access repository that collects the work of Toulouse researchers and makes it freely available over the web where possible. This is an author-deposited version published in : http://oatao.univ- toulouse.fr/ Eprints ID : 11091 To link to this article : doi:10.1002/esp.3515 URL : http://dx.doi.org/10.1002/esp.3515 To cite this version : Corenblit, Dov and Steiger, Johannes and González, Eduardo and Gurnell, Angela M. and Charrier, Gaspard and Darrozes, José and Dousseau, Julien and Julien, Frédéric and Lambs, Luc and Larrue, Sébastien and Roussel, Erwan and Vautier, Franck and Voldoire, Olivier The biogeomorphological life cycle of poplars during the fluvial biogeomorphological succession: a special focus on Populus nigra L. (2014) Earth Surface Processes and Landforms . ISSN 0197- 9337 Any correspondance concerning this service should be sent to the repository administrator: [email protected] DOI: 10.1002/esp.3515 The biogeomorphological life cycle of poplars during the fluvial biogeomorphological succession: a special focus on Populus nigra L. D. Corenblit, 1,2 * J. Steiger, 1,2 E. González, 3 A. M. Gurnell, 4 G. Charrier, 1,2 J. Darrozes, 5 J. Dousseau, 1,2 F. Julien, 6 L. Lambs, 6 S. Larrue, 1,2 E. Roussel, 7 F. Vautier 7 and O. Voldoire 1,2 1 Clermont Université, Maison des Sciences de l ’Homme, 4 rue Ledru, 63057, Clermont-Ferrand Cedex 1, France 2 CNRS, UMR 6042, GEOLAB – Laboratoire de géographie physique et environnementale, 63057, Clermont-Ferrand, -

PDF Hosted at the Radboud Repository of the Radboud University Nijmegen
PDF hosted at the Radboud Repository of the Radboud University Nijmegen The following full text is a publisher's version. For additional information about this publication click this link. http://hdl.handle.net/2066/159489 Please be advised that this information was generated on 2017-12-06 and may be subject to change. UITNODIGING AZOLE RESISTANCE IN AZOLE RESISTANCE AZOLE RESISTANCE IN ASPERGILLUS: voor het bijwonen van de EPIDEMIOLOGY AND SURVEILLANCE openbare verdediging van mijn proefschrift Azole resistance in Aspergillus: epidemiology and surveillance ASPERGILLUS Vrijdag 16 september om 12.30 uur precies U bent van harte welkom : EPIDEMIOLOGY AND SURVEILLANCE | Jan W.M. van der Linden van | Jan W.M. AND SURVEILLANCE : EPIDEMIOLOGY bij deze plechtigheid en de aansluitende receptie in de Aula van de Radboud Universiteit, Comeniuslaan 2 6525 HP Nijmegen Jan van der Linden Kaar 13, 5374 JX, Schaijk [email protected] Paranimfen: Harm van Rooij Jan W.M. van der Linden Elke Appelhof [email protected] AZOLE RESISTANCE IN ASPERGILLUS: EPIDEMIOLOGY AND SURVEILLANCE Jan Wilhelmus Marinus van der Linden Colofon Explanation of the cover: The background photograph is a microscopic view of azole-resistant Aspergillus fumigatus hyphae, recovered from pus from a patient with a kidney transplant. The Aspergillus isolate is projected over a picture of the globe, spreading out from a droplet in the middle of Europe nearby the Netherlands. It represents the global spread and threat of (knowledge and understanding of) resistant Aspergillus fumigatus around the world. ISBN/EAN: 978-90-825729-0-2 Lay-out by Corné van de Laar Cover design by Jan van der Linden The photograph on the cover was used with courtesy of Prof. -

THE KUZNETS CURVE and the LIFE CYCLE ANALOGY by 97/08
THE KUZNETS CURVE AND THE LIFE CYCLE ANALOGY by R. U. AYRES 97/08/EPS This working paper was published in the context of INSEADs Centre for the Management of Environmental Resources, an RD partnership sponsored by Ciba-Geigy, Danfoss, Otto Group and Royal Dutch/Shell and Sandoz AG. * Sandoz Professor of Management and the Environment at INSEAD, Boulevard de Constance, 77305 Fontainebleau Cede; France. A working paper in the INSEAD Working Paper Series is intended as a means whereby a faculty researchers thoughts and findings may be communicated to interested readers. The paper should be considered preliminary in nature and may require revision. Printed at INSEAD, Fontainebleau, France. The Kuznets Curve and the Life Cycle Analogy A Robert U. Ayres Center for the Management of Environmental Resources INSEAD Fontainebleau, France January 1997 Abstract The growth and, especially, the successions of industrial sectors strongly suggests an "aging" phenomenon. In fact, the Kuznets curve was originally based on this notion. More recently, the aging idea has been more fully elaborated into a 'life cycle' scheme (conception, birth, childhood, adolescence, maturity and senescence) which corresponds quite closely with the observed stages of development of a new technology or product. The question arises: is the life cycle analogy sufficiently general to constitute a set of "stylized facts" around which a theory could be developed? In particular, is the life cycle reversible? If so, under what circumstances? The paper addresses these questions and concludes with some observations about the environmental Kuznets curve (EKC) and its applicability to the question of whether economic growth is compatible with long-term sustainability. -

Entomopathogenic Nematodes and Their Bacterial Symbionts: the Inside out of a Mutualistic Association
~ I SYMBIOSIS (2008) 46. 65-75 ©2008 Balaban, Philadelphia/Rehovot ISSN 0334-5114 Review article Entomopathogenic nematodes and their bacterial symbionts: The inside out of a mutualistic association S. Patricia Stock1* and Heidi Goodrich Blair2 'Department of Entomology, University of Arizona, 1140 E. South Campus Dr., Tucson, AZ 85721, USA, Tel. + 1-520-626-3854, Email. [email protected]; 2Department of Bacteriology, University of Wisconsin Madison, 1550 Linden Dr., Madison, WI 53706, USA, Tel.+ 1-608-265-4537, Email. [email protected] (Received October 19, 2007; Accepted January 7, 2008) Abstract Entomopathogenic nematodes Steinernema and Heterorhabditis spp. (Nematoda: Steinemematidae, Heterorhabditidae) and their bacterial symbiont bacteriaXenorhabdus and Photorhabdus spp (Gram-negative Enterobacteriaceae) represent an emerging model of terrestrial animal-microbe symbiotic relationships. Xenorhabdus and Photorhabdus spp. are harbored as symbionts in the intestine of the only free-living stage of the nematodes, also known as the infective juvenile or 3rd stage infective juvenile. The bacterium-nematode pair is pathogenic for a wide range of insects and has successfully been implemented in biological control and integrated pest management programs worldwide. Moreover, realization of the practical use of these nematodes has spurred developments across a far broader scientific front. Recent years have seen an intensive worldwide search for fresh genetic materials resulting in an exponential growth of described new species and the discovery of thousands of new isolates worldwide. These nematodes and their bacterial symbionts are now considered a tractable model system that is amenable to study physiological, chemical, structural and developmental aspects of beneficial symbiotic associations. We herein provide an overview of the research done in relation to the study of the symbiotic interactions between Steinernema and Heterorhabditis nematodes and their bacterial symbionts.