A Study on Four Populations of Tripura, North East India
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Some Principles of the Use of Macro-Areas Language Dynamics &A
Online Appendix for Harald Hammarstr¨om& Mark Donohue (2014) Some Principles of the Use of Macro-Areas Language Dynamics & Change Harald Hammarstr¨om& Mark Donohue The following document lists the languages of the world and their as- signment to the macro-areas described in the main body of the paper as well as the WALS macro-area for languages featured in the WALS 2005 edi- tion. 7160 languages are included, which represent all languages for which we had coordinates available1. Every language is given with its ISO-639-3 code (if it has one) for proper identification. The mapping between WALS languages and ISO-codes was done by using the mapping downloadable from the 2011 online WALS edition2 (because a number of errors in the mapping were corrected for the 2011 edition). 38 WALS languages are not given an ISO-code in the 2011 mapping, 36 of these have been assigned their appropri- ate iso-code based on the sources the WALS lists for the respective language. This was not possible for Tasmanian (WALS-code: tsm) because the WALS mixes data from very different Tasmanian languages and for Kualan (WALS- code: kua) because no source is given. 17 WALS-languages were assigned ISO-codes which have subsequently been retired { these have been assigned their appropriate updated ISO-code. In many cases, a WALS-language is mapped to several ISO-codes. As this has no bearing for the assignment to macro-areas, multiple mappings have been retained. 1There are another couple of hundred languages which are attested but for which our database currently lacks coordinates. -

INTRODUCTION 1 1 Lepcha Is a Tibeto-Burman Language Spoken In
CHAPTER ONE INTRODUCTION 11 Lepcha is a Tibeto-Burman language spoken in Sikkim, Darjeeling district in West Bengal in India, in Ilm district in Nepal, and in a few villages of Samtsi district in south-western Bhutan. The tribal home- land of the Lepcha people is referred to as ne mayLe VÎa ne máyel lyáng ‘hidden paradise’ or ne mayLe malUX VÎa ne máyel málúk lyáng ‘land of eternal purity’. Most of the areas in which Lepcha is spoken today were once Sikkimese territory. The kingdom of Sikkim used to com- prise all of present-day Sikkim and most of Darjeeling district. Kalim- pong, now in Darjeeling district, used to be part of Bhutan, but was lost to the British and became ‘British Bhutan’ before being incorpo- rated into Darjeeling district. The Lepcha are believed to be the abo- riginal inhabitants of Sikkim. Today the Lepcha people constitute a minority of the population of modern Sikkim, which has been flooded by immigrants from Nepal. Although the Lepcha themselves estimate their number of speakers to be over 50,000, the total number is likely to be much smaller. Accord- ing to the 1991 Census of India, the most recent statistical profile for which the data have been disaggregated, the total number of mother tongue Lepcha speakers across the nation is 29,854. While their dis- tribution is largely in Sikkim and the northern districts of West Ben- gal, there are no reliable speaker numbers for these areas. In the Dar- jeeling district there are many Lepcha villages particularly in the area surrounding the small town of Kalimpong. -

Vowel Harmony in Chiru Mechek Sampar Awan, Ph.D
================================================================= Language in India www.languageinindia.com ISSN 1930-2940 Vol. 17:7 July 2017 UGC Approved List of Journals Serial Number 49042 ================================================================ Vowel Harmony in Chiru Mechek Sampar Awan, Ph.D. Scholar ================================================= Abstract This paper is an attempt to describe vowel harmony in Chiru. Chiru is an endangered Kuki-Chin language of Tibeto-Burman language family (Grierson 1904) spoken by 8599 speakers (Census of India: 2011). Harmony is a term used in phonology to refer to the way the articulation of one phonological unit is influenced by another unit in the same word or phrase (Crystal 1997). An analogous notion is that of assimilation. The two main processes are consonant harmony and vowel harmony. Interestingly vowel harmony is found only in a few languages among Kuki-Chin groups, viz., Biate, Chiru etc. Most of the Kuki-Chin languages like Thadou, do not have vowel harmony. The vowel harmony in Chiru mainly occurs in pronominal prefixes and genitives of the first and third persons, causative prefixes, numeral prefixes and a few disyllabic words. This phonological process in Chiru is due to the regressive assimilation of vowels in which the vowel phoneme in the first syllable is influenced by the vowel phoneme in the following syllable to become identical to each other in a word. Keywords: Chiru, Kuki-Chin, Vowel harmony Introduction Chiru, the language spoken by the Chiru people, belongs to the Kuki-Chin group of the Tibeto-Burman language family (Grierson 1904). Chiru is one of the thirty-three recognised tribal languages of Manipur. The total population of Chiru speakers is only 8599 (census of India: 2011). -

7=SINO-INDIAN Phylosector
7= SINO-INDIAN phylosector Observatoire Linguistique Linguasphere Observatory page 525 7=SINO-INDIAN phylosector édition princeps foundation edition DU RÉPERTOIRE DE LA LINGUASPHÈRE 1999-2000 THE LINGUASPHERE REGISTER 1999-2000 publiée en ligne et mise à jour dès novembre 2012 published online & updated from November 2012 This phylosector comprises 22 sets of languages spoken by communities in eastern Asia, from the Himalayas to Manchuria (Heilongjiang), constituting the Sino-Tibetan (or Sino-Indian) continental affinity. See note on nomenclature below. 70= TIBETIC phylozone 71= HIMALAYIC phylozone 72= GARIC phylozone 73= KUKIC phylozone 74= MIRIC phylozone 75= KACHINIC phylozone 76= RUNGIC phylozone 77= IRRAWADDIC phylozone 78= KARENIC phylozone 79= SINITIC phylozone This continental affinity is composed of two major parts: the disparate Tibeto-Burman affinity (zones 70= to 77=), spoken by relatively small communities (with the exception of 77=) in the Himalayas and adjacent regions; and the closely related Chinese languages of the Sinitic set and net (zone 79=), spoken in eastern Asia. The Karen languages of zone 78=, formerly considered part of the Tibeto-Burman grouping, are probably best regarded as a third component of Sino-Tibetan affinity. Zone 79=Sinitic includes the outer-language with the largest number of primary voices in the world, representing the most populous network of contiguous speech-communities at the end of the 20th century ("Mainstream Chinese" or so- called 'Mandarin', standardised under the name of Putonghua). This phylosector is named 7=Sino-Indian (rather than Sino-Tibetan) to maintain the broad geographic nomenclature of all ten sectors of the linguasphere, composed of the names of continental or sub-continental entities. -

Grade 9 Winter Holiday Assignment
[1] Most of the writings, from Sikkim, are mostly in Nepali. The Sikkimese mass never received British mode of education or rather there were no schools that could strictly be called an English medium, barring a few. Most of the scanty English writing by the Sikkimese was mainly restricted to non-fiction books and that too mainly concentrated on the history or polity of Sikkim. The Sikkimese wanted to write their own history first TASK: Students must prepare a presentation in any art form based in the prominent literary figures and literature of sikkim. It could include interview, dramtizing, TV show, a scroll magazine, role play. [2] The numerous monasteries in Gangtok display the finesse of Tibetan Buddhist Architecture. Architectural typologies developed in the Sikkim are in response to climate, and functionality. The materials used are locally available like bamboo, cane, cane leaves, mud, and lime. Nowadays, bricks., stone chips, rock slabs, etc. are also being used. Sloping roofs are a common architecture element in all the typologies because of high rainfall in the region TASK: Students must analyse the architechture of Sikkim .Appreciate its beauty.Analyse the golden ratio. Analyse various polygons /solid figures used in the architechtureConnect it to the chapter SURFACE AREAS AND VOUMES/HERONS FORMULA. The project can be a written project /PPT/.The project should contain: Introduction Description of the architechture ,analyzing solid figures and polygons used Model of architechtures.(Drawing ) Questions connecting to surfaceareas and volumes/Areas related o circles.(create your own questions related to the chapter) https://www.youtube.com/watch?v=x5xjqqx6iok https://www.youtube.com/watch?v=2hWIK1NLAhY [3] Sikkim has abundant hydroelectric power potential and the State Government has taken steps to exploit this potential for revenue generation. -

Language in India
LANGUAGE IN INDIA Strength for Today and Bright Hope for Tomorrow Volume 18:7 July 2018 ISSN 1930-2940 Managing Editor: M. S. Thirumalai, Ph.D. Editors: B. Mallikarjun, Ph.D. Sam Mohanlal, Ph.D. B. A. Sharada, Ph.D. A. R. Fatihi, Ph.D. Lakhan Gusain, Ph.D. Jennifer Marie Bayer, Ph.D. G. Baskaran, Ph.D. L. Ramamoorthy, Ph.D. C. Subburaman, Ph.D. (Economics) N. Nadaraja Pillai, Ph.D. Renuga Devi, Ph.D. Soibam Rebika Devi, M.Sc., Ph.D. Dr. S. Chelliah, M.A., Ph.D. Assistant Managing Editor: Swarna Thirumalai, M.A. Contents Language in India www.languageinindia.com is included in the UGC Approved List of Journals. Serial Number 49042. Materials published in Language in India www.languageinindia.com are indexed in EBSCOHost database, MLA International Bibliography and the Directory of Periodicals, ProQuest (Linguistics and Language Behavior Abstracts) and Gale Research. The journal is included in the Cabell’s Directory, a leading directory in the USA. Articles published in Language in India are peer-reviewed by one or more members of the Board of Editors or an outside scholar who is a specialist in the related field. Since the dissertations are already reviewed by the University-appointed examiners, dissertations accepted for publication in Language in India are not reviewed again. This is our 18th year of publication. All back issues of the journal are accessible through this link: http://languageinindia.com/backissues/2001.html Language in India www.languageinindia.com ISSN 1930-2940 18:7 July 2018 Contents i C.P. Ajitha Sekhar, Ph.D. -

Country Technical Note on Indigenous Peoples' Issues
Country Technical Note on Indigenous Peoples’ Issues Republic of India Country Technical Notes on Indigenous Peoples’ Issues REPUBLIC OF INDIA Submitted by: C.R Bijoy and Tiplut Nongbri Last updated: January 2013 Disclaimer The opinions expressed in this publication are those of the authors and do not necessarily represent those of the International Fund for Agricultural Development (IFAD). The designations employed and the presentation of material in this publication do not imply the expression of any opinion whatsoever on the part of IFAD concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. The designations ‗developed‘ and ‗developing‘ countries are intended for statistical convenience and do not necessarily express a judgment about the stage reached by a particular country or area in the development process. All rights reserved Table of Contents Country Technical Note on Indigenous Peoples‘ Issues – Republic of India ......................... 1 1.1 Definition .......................................................................................................... 1 1.2 The Scheduled Tribes ......................................................................................... 4 2. Status of scheduled tribes ...................................................................................... 9 2.1 Occupation ........................................................................................................ 9 2.2 Poverty .......................................................................................................... -

George Van Driem Lectures Historical Relations Between Sikkim And
George van Driem lectures Historical relations between Sikkim and Nepal since the 17th century and their ethnolinguistic consequences and ramifications, lecture for students and team members of the project « Phonetics, Phonology and New Orthographies: Helping Native Language Communities in the Himalayas », Linguistics Institute, University of Bern, 31 March 2017. Who are the Japanese, and where do the Japanese come from?, invited lecture presented at the workshop ‘Human Evolution in Eurasia elucidated through Genetics, Archaeology and Linguistics’ hosted by the National Institute of Genetics at Mishima, 17 March 2017. Previously unknown episodes in the peopling of the greater Himalayan region, invited lecture at the International Consortium for East Himalayan Ethnolinguistic Prehistory, La Trobe University, Bundoora, 8 February 2017. Transitivity in Lohorung, invited keynote lecture at the Kiranti language workshop, Université Paris Diderot, 2 December 2016. The Denisovan Legacy in Tibet and Beyond, invited guest lecture at the 1st Tibetan Language Linguistic Forum, Nánkāi University, Tiānjīn, 27 August 2016. Some epistemic categories in the Dzongkha verb: The dangers of Platonic essentialism in linguistics and in life, invited guest lecture at the 1st Sino-Tibetan Language Research Methodology Workshop, Nánkāi University, Tiānjīn, 25 August 2016. Proto-Trans-Himalayan verbal morphology: Kiranti languages, the Gongduk and the Black Mountain Mönpa, invited guest lecture at the 1st Sino-Tibetan Language Research Methodology Workshop, Nánkāi University, Tiānjīn, 25 August 2016. Ethnolinguistic phylogeography and prehistory: The Eastern Himalayan region as a cradle of ethnogenesis and linguistic diversification in the prehistoric past, invited guest lecture at the 1st Sino-Tibetan Language Research Methodology Workshop, Nánkāi University, Tiānjīn, 24 August 2016. -

The Naga Language Groups Within the Tibeto-Burman Language Family
TheNaga Language Groups within the Tibeto-Burman Language Family George van Driem The Nagas speak languages of the Tibeto-Burman fami Ethnically, many Tibeto-Burman tribes of the northeast ly. Yet, according to our present state of knowledge, the have been called Naga in the past or have been labelled as >Naga languages< do not constitute a single genetic sub >Naga< in scholarly literature who are no longer usually group within Tibeto-Burman. What defines the Nagas best covered by the modern more restricted sense of the term is perhaps just the label Naga, which was once applied in today. Linguistically, even today's >Naga languages< do discriminately by Indo-Aryan colonists to all scantily clad not represent a single coherent branch of the family, but tribes speaking Tibeto-Burman languages in the northeast constitute several distinct branches of Tibeto-Burman. of the Subcontinent. At any rate, the name Naga, ultimately This essay aims (1) to give an idea of the linguistic position derived from Sanskrit nagna >naked<, originated as a titu of these languages within the family to which they belong, lar label, because the term denoted a sect of Shaivite sadhus (2) to provide a relatively comprehensive list of names and whose most salient trait to the eyes of the lay observer was localities as a directory for consultation by scholars and in that they went through life unclad. The Tibeto-Burman terested laymen who wish to make their way through the tribes labelled N aga in the northeast, though scantily clad, jungle of names and alternative appellations that confront were of course not Hindu at all. -

THE LANGUAGES of MANIPUR: a CASE STUDY of the KUKI-CHIN LANGUAGES* Pauthang Haokip Department of Linguistics, Assam University, Silchar
Linguistics of the Tibeto-Burman Area Volume 34.1 — April 2011 THE LANGUAGES OF MANIPUR: A CASE STUDY OF THE KUKI-CHIN LANGUAGES* Pauthang Haokip Department of Linguistics, Assam University, Silchar Abstract: Manipur is primarily the home of various speakers of Tibeto-Burman languages. Aside from the Tibeto-Burman speakers, there are substantial numbers of Indo-Aryan and Dravidian speakers in different parts of the state who have come here either as traders or as workers. Keeping in view the lack of proper information on the languages of Manipur, this paper presents a brief outline of the languages spoken in the state of Manipur in general and Kuki-Chin languages in particular. The social relationships which different linguistic groups enter into with one another are often political in nature and are seldom based on genetic relationship. Thus, Manipur presents an intriguing area of research in that a researcher can end up making wrong conclusions about the relationships among the various linguistic groups, unless one thoroughly understands which groups of languages are genetically related and distinct from other social or political groupings. To dispel such misconstrued notions which can at times mislead researchers in the study of the languages, this paper provides an insight into the factors linguists must take into consideration before working in Manipur. The data on Kuki-Chin languages are primarily based on my own information as a resident of Churachandpur district, which is further supported by field work conducted in Churachandpur district during the period of 2003-2005 while I was working for the Central Institute of Indian Languages, Mysore, as a research investigator. -
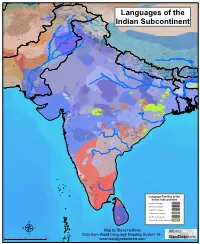
Map by Steve Huffman Data from World Language Mapping System 16
Tajiki Tajiki Tajiki Shughni Southern Pashto Shughni Tajiki Wakhi Wakhi Wakhi Mandarin Chinese Sanglechi-Ishkashimi Sanglechi-Ishkashimi Wakhi Domaaki Sanglechi-Ishkashimi Khowar Khowar Khowar Kati Yidgha Eastern Farsi Munji Kalasha Kati KatiKati Phalura Kalami Indus Kohistani Shina Kati Prasuni Kamviri Dameli Kalami Languages of the Gawar-Bati To rw al i Chilisso Waigali Gawar-Bati Ushojo Kohistani Shina Balti Parachi Ashkun Tregami Gowro Northwest Pashayi Southwest Pashayi Grangali Bateri Ladakhi Northeast Pashayi Southeast Pashayi Shina Purik Shina Brokskat Aimaq Parya Northern Hindko Kashmiri Northern Pashto Purik Hazaragi Ladakhi Indian Subcontinent Changthang Ormuri Gujari Kashmiri Pahari-Potwari Gujari Bhadrawahi Zangskari Southern Hindko Kashmiri Ladakhi Pangwali Churahi Dogri Pattani Gahri Ormuri Chambeali Tinani Bhattiyali Gaddi Kanashi Tinani Southern Pashto Ladakhi Central Pashto Khams Tibetan Kullu Pahari KinnauriBhoti Kinnauri Sunam Majhi Western Panjabi Mandeali Jangshung Tukpa Bilaspuri Chitkuli Kinnauri Mahasu Pahari Eastern Panjabi Panang Jaunsari Western Balochi Southern Pashto Garhwali Khetrani Hazaragi Humla Rawat Central Tibetan Waneci Rawat Brahui Seraiki DarmiyaByangsi ChaudangsiDarmiya Western Balochi Kumaoni Chaudangsi Mugom Dehwari Bagri Nepali Dolpo Haryanvi Jumli Urdu Buksa Lowa Raute Eastern Balochi Tichurong Seke Sholaga Kaike Raji Rana Tharu Sonha Nar Phu ChantyalThakali Seraiki Raji Western Parbate Kham Manangba Tibetan Kathoriya Tharu Tibetan Eastern Parbate Kham Nubri Marwari Ts um Gamale Kham Eastern -

Land, People and Politics: Contest Over Tribal Land in Northeast India
Land, People and Politics Land, PeoPLe and PoLitics: contest oveR tRibaL Land in noRtheast india Editors Walter Fernandes sanjay BarBora North Eastern Social Research Centre International Workgroup for Indigenous Affairs 2008 Land, People and Politics: contest over tribal Land in northeast india Editors: Walter Fernandes and Sanjay Barbora Pages: 178 ISSN: 0105-4503 ISBN: 9788791563409 Language: English Index : 1. Indigenous peoples; 2. Land alienation; Acknowledgements 3. Northeast India; 4. Colonialism Geographical area: Asia Publication date: January 2009 cover design: Kazimuddin Ahmed, Panos South Asia This book is an outcome of collaboration between North Eastern Social Research Centre (NESRC), Panos South Asia and International Published by: North Eastern Social Research Centre 110 Kharghuli Road (1st floor) Work Group for Indigenous Affairs (IWGIA). It is based on studies on Guwahati 781004 land alienation in different states of the Northeast done by a group of Assam, India researchers in 2005-2006. Some papers that were produced during that Tel. (+91-361) 2602819 study are included in this book while others are new and were written Email: [email protected] Website: www.creighton.edu/CollaborativeMinistry/ or revised for this publication. We are grateful to all the researchers for NESRC the hard work they have put into these papers. The study, as well as the book, was funded by the Ministry of Foreign Affairs, Government of International Work Group for Indigenous Affairs (IWGIA) Denmark. The study was coordinated by Artax Shimray. We are grateful Classensgade 11E DK-2100 Copenhagen to the Ministry of Foreign Affairs, Denmark for financial support for this Denmark book. We are grateful to IWGIA particularly Christian Erni and Christina www.iwgia.org Nilsson for their support.