Biophysical and Functional Characterization of Norrin Signaling Through Frizzled4,” by Injin Bang, Hee Ryung Kim, Andrew H
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

G Protein-Coupled Receptors
S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2015/16: G protein-coupled receptors. British Journal of Pharmacology (2015) 172, 5744–5869 THE CONCISE GUIDE TO PHARMACOLOGY 2015/16: G protein-coupled receptors Stephen PH Alexander1, Anthony P Davenport2, Eamonn Kelly3, Neil Marrion3, John A Peters4, Helen E Benson5, Elena Faccenda5, Adam J Pawson5, Joanna L Sharman5, Christopher Southan5, Jamie A Davies5 and CGTP Collaborators 1School of Biomedical Sciences, University of Nottingham Medical School, Nottingham, NG7 2UH, UK, 2Clinical Pharmacology Unit, University of Cambridge, Cambridge, CB2 0QQ, UK, 3School of Physiology and Pharmacology, University of Bristol, Bristol, BS8 1TD, UK, 4Neuroscience Division, Medical Education Institute, Ninewells Hospital and Medical School, University of Dundee, Dundee, DD1 9SY, UK, 5Centre for Integrative Physiology, University of Edinburgh, Edinburgh, EH8 9XD, UK Abstract The Concise Guide to PHARMACOLOGY 2015/16 provides concise overviews of the key properties of over 1750 human drug targets with their pharmacology, plus links to an open access knowledgebase of drug targets and their ligands (www.guidetopharmacology.org), which provides more detailed views of target and ligand properties. The full contents can be found at http://onlinelibrary.wiley.com/doi/ 10.1111/bph.13348/full. G protein-coupled receptors are one of the eight major pharmacological targets into which the Guide is divided, with the others being: ligand-gated ion channels, voltage-gated ion channels, other ion channels, nuclear hormone receptors, catalytic receptors, enzymes and transporters. These are presented with nomenclature guidance and summary information on the best available pharmacological tools, alongside key references and suggestions for further reading. -

Multi-Functionality of Proteins Involved in GPCR and G Protein Signaling: Making Sense of Structure–Function Continuum with In
Cellular and Molecular Life Sciences (2019) 76:4461–4492 https://doi.org/10.1007/s00018-019-03276-1 Cellular andMolecular Life Sciences REVIEW Multi‑functionality of proteins involved in GPCR and G protein signaling: making sense of structure–function continuum with intrinsic disorder‑based proteoforms Alexander V. Fonin1 · April L. Darling2 · Irina M. Kuznetsova1 · Konstantin K. Turoverov1,3 · Vladimir N. Uversky2,4 Received: 5 August 2019 / Revised: 5 August 2019 / Accepted: 12 August 2019 / Published online: 19 August 2019 © Springer Nature Switzerland AG 2019 Abstract GPCR–G protein signaling system recognizes a multitude of extracellular ligands and triggers a variety of intracellular signal- ing cascades in response. In humans, this system includes more than 800 various GPCRs and a large set of heterotrimeric G proteins. Complexity of this system goes far beyond a multitude of pair-wise ligand–GPCR and GPCR–G protein interactions. In fact, one GPCR can recognize more than one extracellular signal and interact with more than one G protein. Furthermore, one ligand can activate more than one GPCR, and multiple GPCRs can couple to the same G protein. This defnes an intricate multifunctionality of this important signaling system. Here, we show that the multifunctionality of GPCR–G protein system represents an illustrative example of the protein structure–function continuum, where structures of the involved proteins represent a complex mosaic of diferently folded regions (foldons, non-foldons, unfoldons, semi-foldons, and inducible foldons). The functionality of resulting highly dynamic conformational ensembles is fne-tuned by various post-translational modifcations and alternative splicing, and such ensembles can undergo dramatic changes at interaction with their specifc partners. -

Wnt/Rspondin/Β-Catenin Signals Control Axonal Sorting and Lineage Progression in Schwann Cell Development
Wnt/Rspondin/β-catenin signals control axonal sorting and lineage progression in Schwann cell development Tamara Grigoryana, Simone Steina, Jingjing Qia, Hagen Wendeb, Alistair N. Garrattc, Klaus-Armin Naved, Carmen Birchmeierb, and Walter Birchmeiera,1 aCancer Research Program and bNeuroscience Program, Max Delbrück Center for Molecular Medicine, 13125 Berlin, Germany; cCenter for Anatomy, Charité University Hospital, 10117 Berlin, Germany; and dDepartment of Neurogenetics, Max Planck Institute for Experimental Medicine, 37075 Göttingen, Germany Edited by Thomas C. Südhof, Stanford University School of Medicine, Stanford, CA, and approved September 26, 2013 (received for review June 2, 2013) During late Schwann cell development, immature Schwann cells organs (24–27). A role of Rspondins and Lgr4–6 receptors in SC segregate large axons from bundles, a process called “axonal ra- development has not been studied. dial sorting.” Here we demonstrate that canonical Wnt signals play Here we define a temporal window of Wnt/β-catenin activity a critical role in radial sorting and assign a role to Wnt and Rspon- and its role in SC lineage progression using mouse genetics and din ligands in this process. Mice carrying β-catenin loss-of-function cell culture techniques. Conditional loss-of-function (LOF) and mutations show a delay in axonal sorting; conversely, gain-of-function gain-of-function (GOF) mutations of β-catenin in mouse SCs mutations result in accelerated sorting. Sorting deficits are accom- produce converse phenotypes: a delay and an acceleration of panied by abnormal process extension, differentiation, and aber- axonal sorting, respectively. Using cultured primary SCs and rant cell cycle exit of the Schwann cells. -

Wnt/Rspondin/Β-Catenin Signals Control Axonal Sorting and Lineage Progression in Schwann Cell Development
Wnt/Rspondin/β-catenin signals control axonal sorting and lineage progression in Schwann cell development Tamara Grigoryana, Simone Steina, Jingjing Qia, Hagen Wendeb, Alistair N. Garrattc, Klaus-Armin Naved, Carmen Birchmeierb, and Walter Birchmeiera,1 aCancer Research Program and bNeuroscience Program, Max Delbrück Center for Molecular Medicine, 13125 Berlin, Germany; cCenter for Anatomy, Charité University Hospital, 10117 Berlin, Germany; and dDepartment of Neurogenetics, Max Planck Institute for Experimental Medicine, 37075 Göttingen, Germany Edited by Thomas C. Südhof, Stanford University School of Medicine, Stanford, CA, and approved September 26, 2013 (received for review June 2, 2013) During late Schwann cell development, immature Schwann cells organs (24–27). A role of Rspondins and Lgr4–6 receptors in SC segregate large axons from bundles, a process called “axonal ra- development has not been studied. dial sorting.” Here we demonstrate that canonical Wnt signals play Here we define a temporal window of Wnt/β-catenin activity a critical role in radial sorting and assign a role to Wnt and Rspon- and its role in SC lineage progression using mouse genetics and din ligands in this process. Mice carrying β-catenin loss-of-function cell culture techniques. Conditional loss-of-function (LOF) and mutations show a delay in axonal sorting; conversely, gain-of-function gain-of-function (GOF) mutations of β-catenin in mouse SCs mutations result in accelerated sorting. Sorting deficits are accom- produce converse phenotypes: a delay and an acceleration of panied by abnormal process extension, differentiation, and aber- axonal sorting, respectively. Using cultured primary SCs and rant cell cycle exit of the Schwann cells. -

WNT Signalling in Chronic Lung Diseases
Thorax Online First, published on April 17, 2017 as 10.1136/thoraxjnl-2016-209753 State of the art review ‘WNT-er is coming’: WNT signalling in chronic lung Thorax: first published as 10.1136/thoraxjnl-2016-209753 on 17 April 2017. Downloaded from diseases H A Baarsma,1 M Königshoff1,2 1Comprehensive Pneumology ABSTRACT involved in the phosphorylation and subsequent Center, Helmholtz Center Chronic lung diseases represent a major public health degradation of β-catenin. Binding of a specific WNT Munich, Ludwig Maximilians University Munich, University problem with only limited therapeutic options. An ligand (eg, WNT-3A) to one of the Frizzled recep- Hospital Grosshadern, Member important unmet need is to identify compounds and tors (FZD1 through FZD10) and subsequent activa- of the German Center for Lung drugs that target key molecular pathways involved in the tion of the low-density lipoprotein receptor-related Research (DZL), Munich, pathogenesis of chronic lung diseases. Over the last proteins 5 and 6 (LRP5/6) co-receptors triggers an Germany 2 decade, there has been extensive interest in investigating intracellular signalling cascade, which results in Division of Pulmonary ‘β ’ Sciences and Critical Care Wingless/integrase-1 (WNT) signalling pathways; and inactivation of the -catenin destruction complex . Medicine, Department of WNT signal alterations have been linked to pulmonary Hence, cytosolic β-catenin can accumulate, translo- Medicine, University of disease pathogenesis and progression. Here, we cate to the nucleus and, in association with T cell Colorado School of Medicine, comprehensively review the cumulative evidence for WNT factor/lymphoid enhancer factor-1 (TCF/LEF) Aurora, Colorado, USA pathway alterations in chronic lung pathologies, including family of transcription factors, induce specific gene 1 Correspondence to idiopathic pulmonary fibrosis, pulmonary arterial expression (figure 1). -

G Protein-Coupled Receptors
Alexander, S. P. H., Christopoulos, A., Davenport, A. P., Kelly, E., Marrion, N. V., Peters, J. A., Faccenda, E., Harding, S. D., Pawson, A. J., Sharman, J. L., Southan, C., Davies, J. A. (2017). THE CONCISE GUIDE TO PHARMACOLOGY 2017/18: G protein-coupled receptors. British Journal of Pharmacology, 174, S17-S129. https://doi.org/10.1111/bph.13878 Publisher's PDF, also known as Version of record License (if available): CC BY Link to published version (if available): 10.1111/bph.13878 Link to publication record in Explore Bristol Research PDF-document This is the final published version of the article (version of record). It first appeared online via Wiley at https://doi.org/10.1111/bph.13878 . Please refer to any applicable terms of use of the publisher. University of Bristol - Explore Bristol Research General rights This document is made available in accordance with publisher policies. Please cite only the published version using the reference above. Full terms of use are available: http://www.bristol.ac.uk/red/research-policy/pure/user-guides/ebr-terms/ S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2017/18: G protein-coupled receptors. British Journal of Pharmacology (2017) 174, S17–S129 THE CONCISE GUIDE TO PHARMACOLOGY 2017/18: G protein-coupled receptors Stephen PH Alexander1, Arthur Christopoulos2, Anthony P Davenport3, Eamonn Kelly4, Neil V Marrion4, John A Peters5, Elena Faccenda6, Simon D Harding6,AdamJPawson6, Joanna L Sharman6, Christopher Southan6, Jamie A Davies6 and CGTP Collaborators 1 School of Life Sciences, -

Adenylyl Cyclase 2 Selectively Regulates IL-6 Expression in Human Bronchial Smooth Muscle Cells Amy Sue Bogard University of Tennessee Health Science Center
University of Tennessee Health Science Center UTHSC Digital Commons Theses and Dissertations (ETD) College of Graduate Health Sciences 12-2013 Adenylyl Cyclase 2 Selectively Regulates IL-6 Expression in Human Bronchial Smooth Muscle Cells Amy Sue Bogard University of Tennessee Health Science Center Follow this and additional works at: https://dc.uthsc.edu/dissertations Part of the Medical Cell Biology Commons, and the Medical Molecular Biology Commons Recommended Citation Bogard, Amy Sue , "Adenylyl Cyclase 2 Selectively Regulates IL-6 Expression in Human Bronchial Smooth Muscle Cells" (2013). Theses and Dissertations (ETD). Paper 330. http://dx.doi.org/10.21007/etd.cghs.2013.0029. This Dissertation is brought to you for free and open access by the College of Graduate Health Sciences at UTHSC Digital Commons. It has been accepted for inclusion in Theses and Dissertations (ETD) by an authorized administrator of UTHSC Digital Commons. For more information, please contact [email protected]. Adenylyl Cyclase 2 Selectively Regulates IL-6 Expression in Human Bronchial Smooth Muscle Cells Document Type Dissertation Degree Name Doctor of Philosophy (PhD) Program Biomedical Sciences Track Molecular Therapeutics and Cell Signaling Research Advisor Rennolds Ostrom, Ph.D. Committee Elizabeth Fitzpatrick, Ph.D. Edwards Park, Ph.D. Steven Tavalin, Ph.D. Christopher Waters, Ph.D. DOI 10.21007/etd.cghs.2013.0029 Comments Six month embargo expired June 2014 This dissertation is available at UTHSC Digital Commons: https://dc.uthsc.edu/dissertations/330 Adenylyl Cyclase 2 Selectively Regulates IL-6 Expression in Human Bronchial Smooth Muscle Cells A Dissertation Presented for The Graduate Studies Council The University of Tennessee Health Science Center In Partial Fulfillment Of the Requirements for the Degree Doctor of Philosophy From The University of Tennessee By Amy Sue Bogard December 2013 Copyright © 2013 by Amy Sue Bogard. -

Topological Control of Cytokine Receptor Signaling Induces Differential Effects in Hematopoiesis Kritika Mohan, George Ueda, Ah Ram Kim, Kevin M
RESEARCH ◥ control the relative orientation of the ECDs in RESEARCH ARTICLE SUMMARY the dimeric complex. The “angle” series varied the scissor angle between the two ECDs, whereas the “distance” series varied their relative proxim- PROTEIN ENGINEERING ity. The designed DARPin ligands were validated by means of x-ray crystallography for representa- tive complexes. The systematic variation of angular Topological control of cytokine and distance parameters generated a range of full, biased, and partial agonism of EpoR signaling receptor signaling induces differential ◥ in the human erythroid cell ON OUR WEBSITE line UT7/EPO, as shown with flow-cytometry and effects in hematopoiesis Read the full article at http://dx.doi. immunoblotting for phos- Kritika Mohan*, George Ueda*, Ah Ram Kim†, Kevin M. Jude†, Jorge A. Fallas†, org/10.1126/ phorylated downstream science.aav7532 effectors. In general, in- Yu Guo†, Maximillian Hafer, Yi Miao, Robert A. Saxton, Jacob Piehler, .................................................. Vijay G. Sankaran, David Baker‡, K. Christopher Garcia‡ creasing the angle or dis- tance between the receptor ECDs resulted in a progressive partial agonism, as measured with INTRODUCTION: Receptor dimerization is a system, a well-characterized dimeric cytokine changes in maximum response achieved (Emax) fundamental mechanism by which most cyto- receptor system. and median effective concentration (EC50). Biased kines and growth factors activate Type-I trans- signal transducer and activator of transcription Downloaded from membrane receptors. Although previous studies RESULTS: We used the DARPin (designed (STAT) activation was elicited by some of the sur- have shown that ligand-induced topological ankyrin repeat protein) scaffold because of rogate DARPin ligands. We also evaluated the changes in the extracellular domains effects of these DARPin agonists on dif- (ECDs) of dimeric receptors can affect ferentiation and proliferation of hemato- signaling output, the physiological poietic stem and progenitor cells (HSPCs) relevance is not well understood. -
List of Predicted Circfam120a Target Mrnas Gene in Pathway Species Gene Name
List of predicted circFAM120A target mRNAs Gene in pathway Species Gene name HRAS Homo sapiens HRas proto-oncogene, GTPase (HRAS) ADCY1 Homo sapiens Adenylate cyclase 1 (ADCY1) ADCY2 Homo sapiens Adenylate cyclase 2 (ADCY2) ADCY7 Homo sapiens Adenylate cyclase 7 (ADCY7) PTGS2 Homo sapiens Prostaglandin-endoperoxide synthase 2 (PTGS2) PGF Homo sapiens Placental growth factor (PGF) ADCY5 Homo sapiens Adenylate cyclase 5 (ADCY5) STAT5A Homo sapiens Signal transducer and activator of transcription 5A (STAT5A) ADCY6 Homo sapiens Adenylate cyclase 6 (ADCY6) STAT5B Homo sapiens Signal transducer and activator of transcription 5B (STAT5B) LPAR3 Homo sapiens Lysophosphatidic acid receptor 3 (LPAR3) LPAR2 Homo sapiens Lysophosphatidic acid receptor 2 (LPAR2) CTNNB1 Homo sapiens Catenin beta 1 (CTNNB1) CUL2 Homo sapiens Cullin 2 (CUL2) RARA Homo sapiens Retinoic acid receptor alpha (RARA) GNG2 Homo sapiens G protein subunit gamma 2 (GNG2) RARB Homo sapiens Retinoic acid receptor beta (RARB) GNG3 Homo sapiens G protein subunit gamma 3 (GNG3) FAS Homo sapiens Fas cell surface death receptor (FAS) GNG4 Homo sapiens G protein subunit gamma 4 (GNG4) GNG7 Homo sapiens G protein subunit gamma 7 (GNG7) PIK3CG Homo sapiens Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma (PIK3CG) WNT10B Homo sapiens Wnt family member 10B (WNT10B) BCR Homo sapiens BCR, RhoGEF and GTPase activating protein (BCR) BRAF Homo sapiens B-Raf proto-oncogene, serine/threonine kinase (BRAF) RXRB Homo sapiens Retinoid X receptor beta (RXRB) ROCK2 Homo sapiens Rho -

The WNT Receptor FZD7 Is Required for Maintenance of the Pluripotent State in Human Embryonic Stem Cells
The WNT receptor FZD7 is required for maintenance of the pluripotent state in human embryonic stem cells Antonio Fernandeza,1, Ian J. Hugginsa,1, Luca Pernaa, David Brafmana, Desheng Lub,2, Shiyin Yaob, Terry Gaasterlandc, Dennis A. Carsonb,3, and Karl Willerta,3 aDepartment of Cellular and Molecular Medicine, Stem Cell Program, University of California, San Diego, La Jolla, CA 92093-0695; bRebecca and John Moores Cancer Center, Sanford Consortium for Regenerative Medicine, University of California, San Diego, La Jolla, CA 92093; and cScripps Institution of Oceanography and Institute for Genomic Medicine, University of California, San Diego, La Jolla, CA 92093-0202 Contributed by Dennis A. Carson, December 20, 2013 (sent for review October 15, 2013) WNT signaling is involved in maintaining stem cells in an un- relative expression levels of all 10 FZD genes in hESCs using differentiated state; however, it is often unclear which WNTs and a whole-transcriptome sequencing (RNA-seq) data set. This anal- WNT receptors are mediating these activities. Here we examined ysis demonstrated that FZD7 is the most abundantly expressed the role of the WNT receptor FZD7 in maintaining human embry- FZD gene in the hESC line H1/WA01 (Fig. 1A). Levels of the onic stem cells (hESCs) in an undifferentiated and pluripotent second and third most highly expressed FZD genes, FZD5 and state. FZD7 expression is significantly elevated in undifferentiated FZD3, were 4.2-fold and 8.8-fold lower, respectively. That FZD7 is cells relative to differentiated cell populations, and interfering with the most abundantly expressed FZD gene was confirmed using its expression or function, either by short hairpin RNA-mediated quantitative reverse transcription PCR (qRT-PCR) in a separate knockdown or with a fragment antigen binding (Fab) molecule di- hESC line, HUES9 (Fig. -
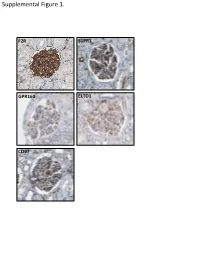
Supplemental Data
Supplemental Figure 1. F2R S1PR1 GPR160 ELTD1 CD97 Supplemental Figure 2. A B brain heart lung liver kidney spleen testis skm Expression in mouse ssues glom rok Gprc5a Gapdh Human Protein Atlas RNAseq database Supplemental Figure 3. A Gprc5a Pdgfrb Merged CL CL Gprc5a CD31 Merged CL CL B C 1.4 * 1.2 mc 1 matrix 0.8 0.6 0.4 matrix 0.2 0 pod end 1 2 mes3 D Vector Gprc5a E 40 kD - - Gprc5a - acn Supplemental Figure 4. Control 12 month-old KO 12 month-old A B C D E F 1 2 0.8 * 1.5 score 0.6 m µ 1 0.4 0.2 Slits/ 0.5 Mesangial 0 0 Ctrl KO Ctrl KO Supplemental Figure 5. A B 1.2 Vector Gprc5a 1 * 0.8 0.6 * 0.4 * Normalized Density 0.2 0 pEGFR/tEGR pSmad/tSmad TGF-β1 C 1.8 siCON 1.6 siGprc5a * 1.4 * * 1.2 1 0.8 0.6 NormalizedDensity 0.4 0.2 0 pEGFR/tEGR pSmad/tSmad TGF-β1 Supplemental table 1. List of glomerulus-expressed GPCRs as detected by qPCR. Data shown as mean ± standard deviation (Glom=glomerulus, Rok=rest of kidney). GPCR Glom Rok Glom/Rok LPAR6 41892,11 ± 38478,89 1040,06 ± 1370,12 39,28 ELTD1 30275,64 ± 14085,26 33,23 ± 46,99 910,13 GPR116 24020,06 ± 7789,84 55,1 ± 47,75 434,96 PTH1R 15402,81 ± 17644,32 7521,01 ± 3264,57 1,05 CALCRL 14096,09 ± 3854,84 199,06 ± 222,52 69,81 HPRT1 13342,04 ± 10677,69 1824,77 ± 1767,23 6,31 S1PR5 9474,7 ± 10124,3 110,84 ± 29,54 84,48 LPHN2 8645,89 ± 914,74 256,04 ± 293,87 32,77 FZD1 8176,2 ± 4947,45 1321,97 ± 1311,91 5,18 CXCR4 7097,31 ± 4388,91 535,98 ± 640,28 12,24 GPR160 6446,59 ± 1550,07 4816,3 ± 5918,99 0,34 NPY1R 6177,74 ± 6282,76 209,64 ± 296,48 28,47 PTGER4 5323,66 ± 3789,51 179,13 ± 210 28,72 RXFP1