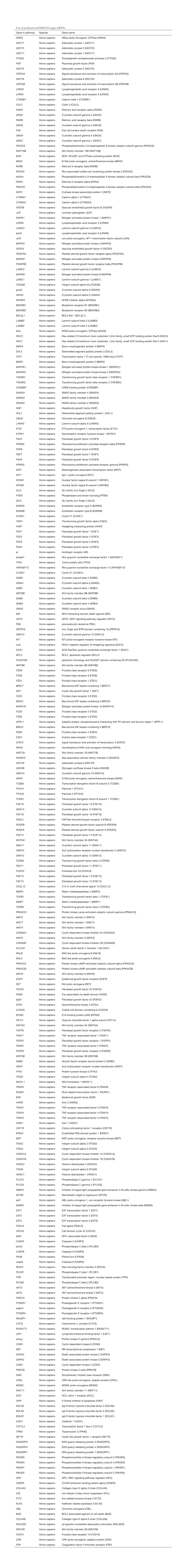
List of Predicted Circfam120a Target Mrnas Gene in Pathway Species Gene Name
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Endothelin System and Therapeutic Application of Endothelin Receptor
xperim ACCESS Freely available online & E en OPEN l ta a l ic P in h l a C r m f o a c l a o n l o r g u y o J Journal of ISSN: 2161-1459 Clinical & Experimental Pharmacology Research Article Endothelin System and Therapeutic Application of Endothelin Receptor Antagonists Abebe Basazn Mekuria, Zemene Demelash Kifle*, Mohammedbrhan Abdelwuhab Department of Pharmacology, School of Pharmacy, College of Medicine and Health Sciences, University of Gondar, Gondar, Ethiopia ABSTRACT Endothelin is a 21 amino acid molecule endogenous potent vasoconstrictor peptide. Endothelin is synthesized in vascular endothelial and smooth muscle cells, as well as in neural, renal, pulmonic, and inflammatory cells. It acts through a seven transmembrane endothelin receptor A (ETA) and endothelin receptor B (ETB) receptors belongs to G protein-coupled rhodopsin-type receptor superfamily. This peptide involved in pathogenesis of cardiovascular disorder like (heart failure, arterial hypertension, myocardial infraction and atherosclerosis), renal failure, pulmonary arterial hypertension and it also involved in pathogenesis of cancer. Potentially endothelin receptor antagonist helps the treatment of the above disorder. Currently, there are a lot of trails both per-clinical and clinical on endothelin antagonist for various cardiovascular, pulmonary and cancer disorder. Some are approved by FAD for the treatment. These agents are including both selective and non-selective endothelin receptor antagonist (ETA/B). Currently, Bosentan, Ambrisentan, and Macitentan approved -

Fog2 Is Critical for Cardiac Function and Maintenance of Coronary Vasculature in the Adult Mouse Heart
Fog2 is critical for cardiac function and maintenance of coronary vasculature in the adult mouse heart Bin Zhou, … , Sergei G. Tevosian, William T. Pu J Clin Invest. 2009;119(6):1462-1476. https://doi.org/10.1172/JCI38723. Research Article Cardiology Aberrant transcriptional regulation contributes to the pathogenesis of both congenital and adult forms of heart disease. While the transcriptional regulator friend of Gata 2 (FOG2) is known to be essential for heart morphogenesis and coronary development, its tissue-specific function has not been previously investigated. Additionally, little is known about the role of FOG2 in the adult heart. Here we used spatiotemporally regulated inactivation of Fog2 to delineate its function in both the embryonic and adult mouse heart. Early cardiomyocyte-restricted loss of Fog2 recapitulated the cardiac and coronary defects of the Fog2 germline murine knockouts. Later cardiomyocyte-restricted loss ofF og2 (Fog2MC) did not result in defects in cardiac structure or coronary vessel formation. However, Fog2MC adult mice had severely depressed ventricular function and died at 8–14 weeks. Fog2MC adult hearts displayed a paucity of coronary vessels, associated with myocardial hypoxia, increased cardiomyocyte apoptosis, and cardiac fibrosis. Induced inactivation of Fog2 in the adult mouse heart resulted in similar phenotypes, as did ablation of the FOG2 interaction with the transcription factor GATA4. Loss of the FOG2 or FOG2-GATA4 interaction altered the expression of a panel of angiogenesis-related genes. Collectively, our data indicate that FOG2 regulates adult heart function and coronary angiogenesis. Find the latest version: https://jci.me/38723/pdf Research article Fog2 is critical for cardiac function and maintenance of coronary vasculature in the adult mouse heart Bin Zhou,1,2 Qing Ma,1 Sek Won Kong,1 Yongwu Hu,1,3 Patrick H. -

F2RL2 Antibody Cat
F2RL2 Antibody Cat. No.: 56-323 F2RL2 Antibody F2RL2 Antibody immunohistochemistry analysis in formalin fixed and paraffin embedded human heart tissue followed by peroxidase conjugation of the secondary antibody and DAB staining. Specifications HOST SPECIES: Rabbit SPECIES REACTIVITY: Human This F2RL2 antibody is generated from rabbits immunized with a KLH conjugated IMMUNOGEN: synthetic peptide between 21-50 amino acids from the N-terminal region of human F2RL2. TESTED APPLICATIONS: IHC-P, WB For WB starting dilution is: 1:1000 APPLICATIONS: For IHC-P starting dilution is: 1:10~50 PREDICTED MOLECULAR 43 kDa WEIGHT: September 25, 2021 1 https://www.prosci-inc.com/f2rl2-antibody-56-323.html Properties This antibody is purified through a protein A column, followed by peptide affinity PURIFICATION: purification. CLONALITY: Polyclonal ISOTYPE: Rabbit Ig CONJUGATE: Unconjugated PHYSICAL STATE: Liquid BUFFER: Supplied in PBS with 0.09% (W/V) sodium azide. CONCENTRATION: batch dependent Store at 4˚C for three months and -20˚C, stable for up to one year. As with all antibodies STORAGE CONDITIONS: care should be taken to avoid repeated freeze thaw cycles. Antibodies should not be exposed to prolonged high temperatures. Additional Info OFFICIAL SYMBOL: F2RL2 Proteinase-activated receptor 3, PAR-3, Coagulation factor II receptor-like 2, Thrombin ALTERNATE NAMES: receptor-like 2, F2RL2, PAR3 ACCESSION NO.: O00254 GENE ID: 2151 USER NOTE: Optimal dilutions for each application to be determined by the researcher. Background and References Coagulation factor II (thrombin) receptor-like 2 (F2RL2) is a member of the large family of 7-transmembrane-region receptors that couple to guanosine-nucleotide-binding proteins. -

Lysophosphatidic Acid and Its Receptors: Pharmacology and Therapeutic Potential in Atherosclerosis and Vascular Disease
JPT-107404; No of Pages 13 Pharmacology & Therapeutics xxx (2019) xxx Contents lists available at ScienceDirect Pharmacology & Therapeutics journal homepage: www.elsevier.com/locate/pharmthera Lysophosphatidic acid and its receptors: pharmacology and therapeutic potential in atherosclerosis and vascular disease Ying Zhou a, Peter J. Little a,b, Hang T. Ta a,c, Suowen Xu d, Danielle Kamato a,b,⁎ a School of Pharmacy, University of Queensland, Pharmacy Australia Centre of Excellence, Woolloongabba, QLD 4102, Australia b Department of Pharmacy, Xinhua College of Sun Yat-sen University, Tianhe District, Guangzhou 510520, China c Australian Institute for Bioengineering and Nanotechnology, The University of Queensland, Brisbane, St Lucia, QLD 4072, Australia d Aab Cardiovascular Research Institute, Department of Medicine, University of Rochester School of Medicine and Dentistry, Rochester, NY 14642, USA article info abstract Available online xxxx Lysophosphatidic acid (LPA) is a collective name for a set of bioactive lipid species. Via six widely distributed G protein-coupled receptors (GPCRs), LPA elicits a plethora of biological responses, contributing to inflammation, Keywords: thrombosis and atherosclerosis. There have recently been considerable advances in GPCR signaling especially Lysophosphatidic acid recognition of the extended role for GPCR transactivation of tyrosine and serine/threonine kinase growth factor G-protein coupled receptors receptors. This review covers LPA signaling pathways in the light of new information. The use of transgenic and Atherosclerosis gene knockout animals, gene manipulated cells, pharmacological LPA receptor agonists and antagonists have Gproteins fi β-arrestins provided many insights into the biological signi cance of LPA and individual LPA receptors in the progression Transactivation of atherosclerosis and vascular diseases. -

ARTICLES Fibroblast Growth Factors 1, 2, 17, and 19 Are The
0031-3998/07/6103-0267 PEDIATRIC RESEARCH Vol. 61, No. 3, 2007 Copyright © 2007 International Pediatric Research Foundation, Inc. Printed in U.S.A. ARTICLES Fibroblast Growth Factors 1, 2, 17, and 19 Are the Predominant FGF Ligands Expressed in Human Fetal Growth Plate Cartilage PAVEL KREJCI, DEBORAH KRAKOW, PERTCHOUI B. MEKIKIAN, AND WILLIAM R. WILCOX Medical Genetics Institute [P.K., D.K., P.B.M., W.R.W.], Cedars-Sinai Medical Center, Los Angeles, California 90048; Department of Obstetrics and Gynecology [D.K.] and Department of Pediatrics [W.R.W.], UCLA School of Medicine, Los Angeles, California 90095 ABSTRACT: Fibroblast growth factors (FGF) regulate bone growth, (G380R) or TD (K650E) mutations (4–6). When expressed at but their expression in human cartilage is unclear. Here, we deter- physiologic levels, FGFR3-G380R required, like its wild-type mined the expression of entire FGF family in human fetal growth counterpart, ligand for activation (7). Similarly, in vitro cul- plate cartilage. Using reverse transcriptase PCR, the transcripts for tivated human TD chondrocytes as well as chondrocytes FGF1, 2, 5, 8–14, 16–19, and 21 were found. However, only FGF1, isolated from Fgfr3-K644M mice had an identical time course 2, 17, and 19 were detectable at the protein level. By immunohisto- of Fgfr3 activation compared with wild-type chondrocytes and chemistry, FGF17 and 19 were uniformly expressed within the showed no receptor activation in the absence of ligand (8,9). growth plate. In contrast, FGF1 was found only in proliferating and hypertrophic chondrocytes whereas FGF2 localized predominantly to Despite the importance of the FGF ligand for activation of the resting and proliferating cartilage. -

Disruption of Fibroblast Growth Factor Signal
Cancer Therapy: Preclinical Disruption of Fibroblast Growth Factor Signal Pathway Inhibits the Growth of Synovial Sarcomas: Potential Application of Signal Inhibitors to MolecularTarget Therapy Ta t s u y a I s hi b e , 1, 2 Tomitaka Nakayama,2 Ta k e s h i O k a m o t o, 1, 2 Tomoki Aoyama,1Koichi Nishijo,1, 2 Kotaro Roberts Shibata,1, 2 Ya s u ko Shim a ,1, 2 Satoshi Nagayama,3 Toyomasa Katagiri,4 Yusuke Nakamura, 4 Takashi Nakamura,2 andJunya Toguchida 1 Abstract Purpose: Synovial sarcoma is a soft tissue sarcoma, the growth regulatory mechanisms of which are unknown.We investigatedthe involvement of fibroblast growth factor (FGF) signals in synovial sarcoma andevaluatedthe therapeutic effect of inhibiting the FGF signal. Experimental Design:The expression of 22 FGF and4 FGF receptor (FGFR) genes in18prima- ry tumors andfive cell lines of synovial sarcoma were analyzedby reverse transcription-PCR. Effects of recombinant FGF2, FGF8, andFGF18 for the activation of mitogen-activatedprotein kinase (MAPK) andthe growth of synovial sarcoma cell lines were analyzed.Growth inhibitory effects of FGFR inhibitors on synovial sarcoma cell lines were investigated in vitro and in vivo. Results: Synovial sarcoma cell lines expressedmultiple FGF genes especially those expressed in neural tissues, among which FGF8 showedgrowth stimulatory effects in all synovial sarcoma cell lines. FGF signals in synovial sarcoma induced the phosphorylation of extracellular signal ^ regulatedkinase (ERK1/2) andp38MAPK but not c-Jun NH 2-terminal kinase. Disruption of the FGF signaling pathway in synovial sarcoma by specific inhibitors of FGFR causedcell cycle ar- rest leading to significant growth inhibition both in vitro and in vivo.Growthinhibitionbythe FGFR inhibitor was associatedwith a down-regulation of phosphorylatedERK1/2 but not p38MAPK, andan ERK kinase inhibitor also showedgrowth inhibitory effects for synovial sar- coma, indicating that the growth stimulatory effect of FGF was transmitted through the ERK1/2. -

FGF14 Regulates Presynaptic Ca2+ Channels and Synaptic Transmission
Cell Reports Article FGF14 Regulates Presynaptic Ca2+ Channels and Synaptic Transmission Haidun Yan,1,3 Juan L. Pablo,2,3 and Geoffrey S. Pitt1,2,3,* 1Division of Cardiology, Department of Medicine, Duke University Medical Center, Durham, NC 27710, USA 2Department of Neurobiology, Duke University Medical Center, Durham, NC 27710, USA 3Ion Channel Research Unit, Duke University Medical Center, Durham, NC 27710, USA *Correspondence: [email protected] http://dx.doi.org/10.1016/j.celrep.2013.06.012 This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited. SUMMARY data pinpointed FGF14 as the locus for spinocerebellar ataxia 27 (SCA27). Fibroblast growth factor homologous factors (FHFs) Focus on FHF regulation of neuronal excitability began when are not growth factors, but instead bind to voltage- Fgf14–/– mice showed ataxia (Wang et al., 2002), providing + gated Na channels (NaV) and regulate their function. a basis for exploring the implications of a linkage analysis that Mutations in FGF14, an FHF that is the locus for identified a F150S missense mutation in a ‘‘b’’ splice variant of F150S F145S spinocerebellar ataxia 27 (SCA27), are believed to FGF14 (FGF14b ; termed FGF14 in some studies that be pathogenic because of a dominant-negative used numbering based on the alternatively spliced FGF14a variant) as the etiology of the autosomal-dominant SCA27 in reduction of Na currents in cerebellar granule cells. V an extended Dutch family (van Swieten et al., 2003). -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Supplementary Table 3 Complete List of RNA-Sequencing Analysis of Gene Expression Changed by ≥ Tenfold Between Xenograft and Cells Cultured in 10%O2
Supplementary Table 3 Complete list of RNA-Sequencing analysis of gene expression changed by ≥ tenfold between xenograft and cells cultured in 10%O2 Expr Log2 Ratio Symbol Entrez Gene Name (culture/xenograft) -7.182 PGM5 phosphoglucomutase 5 -6.883 GPBAR1 G protein-coupled bile acid receptor 1 -6.683 CPVL carboxypeptidase, vitellogenic like -6.398 MTMR9LP myotubularin related protein 9-like, pseudogene -6.131 SCN7A sodium voltage-gated channel alpha subunit 7 -6.115 POPDC2 popeye domain containing 2 -6.014 LGI1 leucine rich glioma inactivated 1 -5.86 SCN1A sodium voltage-gated channel alpha subunit 1 -5.713 C6 complement C6 -5.365 ANGPTL1 angiopoietin like 1 -5.327 TNN tenascin N -5.228 DHRS2 dehydrogenase/reductase 2 leucine rich repeat and fibronectin type III domain -5.115 LRFN2 containing 2 -5.076 FOXO6 forkhead box O6 -5.035 ETNPPL ethanolamine-phosphate phospho-lyase -4.993 MYO15A myosin XVA -4.972 IGF1 insulin like growth factor 1 -4.956 DLG2 discs large MAGUK scaffold protein 2 -4.86 SCML4 sex comb on midleg like 4 (Drosophila) Src homology 2 domain containing transforming -4.816 SHD protein D -4.764 PLP1 proteolipid protein 1 -4.764 TSPAN32 tetraspanin 32 -4.713 N4BP3 NEDD4 binding protein 3 -4.705 MYOC myocilin -4.646 CLEC3B C-type lectin domain family 3 member B -4.646 C7 complement C7 -4.62 TGM2 transglutaminase 2 -4.562 COL9A1 collagen type IX alpha 1 chain -4.55 SOSTDC1 sclerostin domain containing 1 -4.55 OGN osteoglycin -4.505 DAPL1 death associated protein like 1 -4.491 C10orf105 chromosome 10 open reading frame 105 -4.491 -

Bradykinin Receptor Deficiency Or Antagonism Do Not Impact the Host
Ding et al. Intensive Care Medicine Experimental (2019) 7:14 Intensive Care Medicine https://doi.org/10.1186/s40635-019-0228-3 Experimental RESEARCH Open Access Bradykinin receptor deficiency or antagonism do not impact the host response during gram-negative pneumonia-derived sepsis Chao Ding1,2, Jack Yang2, Cornelis van’t Veer2 and Tom van der Poll2,3* * Correspondence: t.vanderpoll@ amc.uva.nl Abstract 2Center of Experimental and Molecular Medicine, Academic Background: Kinins are short peptides with a wide range of proinflammatory Medical Center, University of properties that are generated from kininogens in the so-called kallikrein-kinin system. Amsterdam, Meibergdreef 9, Room Kinins exert their biological activities through stimulation of two distinct receptor G2-130, 1105 AZ Amsterdam, the Netherlands subtypes, the kinin or bradykinin B1 and B2 receptors (B1R, B2R). Acute challenge 3Division of Infectious Diseases, models have implicated B1R and B2R in the pathogenesis of sepsis. However, their Academic Medical Center, role in the host response during sepsis originating from the lung is not known. University of Amsterdam, Amsterdam, the Netherlands Results: To determine the role of B1R and B2R in pneumonia-derived sepsis, B1R/ Full list of author information is B2R-deficient mice and wild-type mice treated with the B1R antagonist R-715 or the available at the end of the article B2R antagonist HOE-140 were studied after infection with the common gram- negative pathogen Klebsiella pneumoniae via the airways. Neither B1R/B2R deficiency nor B1R or B2R inhibition influenced bacterial growth at the primary site of infection or dissemination to distant body sites. -

An Explanation of G-Protein Coupled Receptor Structure
Journal of Stem Cell Research & Therapeutics Review Article Open Access Role of GPCRS towards cell: an explanation of g-protein coupled receptor structure Abstract Volume 2 Issue 3 - 2017 G- protein coupled receptors are the heptahelical, serpentine receptor and are Nidhi Sharma,1 Anil Kumar Sahdev,2 Vinit Raj2 multifunctional receptors having modulatory activity of a wide variety of biological 1Dr. K. N. Modi Institute of Pharmacy & Research centre, process including: Neurotransmission, Chemoattraction, Cardiac function, Olfaction Modinagar, India. and Vision etc. At largest level they are involved in signal transduction across cell 2Department of Pharmaceutical Sciences, Babasaheb Bhimrao membranes therefore they represent major targets in the development of novel drug Ambedkar University, India candidates in all clinical areas. Particularly membrane cholesterol has been reported to have a great role in the functioning of a number of GPCRs. Apart from this it also Correspondence: Vinit Raj, Department of Pharmaceutical have some drawbacks. Mutations that occur are associated with a broad spectrum of Sciences, Babasaheb Bhimrao Ambedkar University, VidyaVihar, diseases of diverse etiology. As a mutations result, there is a change in receptor activity Rae Bareli Road, Lucknow-226025, Email [email protected] (GPCR become inactive, overactive, or constitutively active), in the process of ligand binding and signal transduction. Changes in the GPCRs functioning can cause diseases Received: January 31, 2017 | Published: March 27, 2017 such as retinitis pigmentosa (rhodopsin mutations), nephrogenic diabetes insipidus (vasopressin receptor mutations), and obesity (melanocortin receptor mutations). Keywords: gpcrs, serpentine receptor, neurotransmission, retinitis pigmentosa, nephrogenic diabetes insipidus, bardet-biedl syndrome, hypogonadism Introduction deliver a message and appropriate cellular and physiological response1,2 (Figure 1). -

Instability Restricts Signaling of Multiple Fibroblast Growth Factors
Cell. Mol. Life Sci. DOI 10.1007/s00018-015-1856-8 Cellular and Molecular Life Sciences RESEARCH ARTICLE Instability restricts signaling of multiple fibroblast growth factors Marcela Buchtova • Radka Chaloupkova • Malgorzata Zakrzewska • Iva Vesela • Petra Cela • Jana Barathova • Iva Gudernova • Renata Zajickova • Lukas Trantirek • Jorge Martin • Michal Kostas • Jacek Otlewski • Jiri Damborsky • Alois Kozubik • Antoni Wiedlocha • Pavel Krejci Received: 18 June 2014 / Revised: 7 February 2015 / Accepted: 9 February 2015 Ó Springer Basel 2015 Abstract Fibroblast growth factors (FGFs) deliver ex- failure to activate FGF receptor signal transduction over tracellular signals that govern many developmental and long periods of time, and influence specific cell behavior regenerative processes, but the mechanisms regulating FGF in vitro and in vivo. Stabilization via exogenous heparin signaling remain incompletely understood. Here, we ex- binding, introduction of stabilizing mutations or lowering plored the relationship between intrinsic stability of FGF the cell cultivation temperature rescues signaling of un- proteins and their biological activity for all 18 members of stable FGFs. Thus, the intrinsic ligand instability is an the FGF family. We report that FGF1, FGF3, FGF4, FGF6, important elementary level of regulation in the FGF sig- FGF8, FGF9, FGF10, FGF16, FGF17, FGF18, FGF20, and naling system. FGF22 exist as unstable proteins, which are rapidly de- graded in cell cultivation media. Biological activity of Keywords Fibroblast growth factor Á FGF Á Unstable Á FGF1, FGF3, FGF4, FGF6, FGF8, FGF10, FGF16, FGF17, Proteoglycan Á Regulation and FGF20 is limited by their instability, manifesting as Electronic supplementary material The online version of this article (doi:10.1007/s00018-015-1856-8) contains supplementary material, which is available to authorized users.