Human G Protein Coupled Receptors 384HT
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Activation of the Glucagon-Like Peptide-1 (GLP-1) Receptor by Peptide and Non-Peptide Ligands
The Activation of the Glucagon-Like Peptide-1 (GLP-1) Receptor by Peptide and Non-Peptide Ligands Clare Louise Wishart Submitted in accordance with the requirements for the degree of Doctor of Philosophy of Science University of Leeds School of Biomedical Sciences Faculty of Biological Sciences September 2013 I Intellectual Property and Publication Statements The candidate confirms that the work submitted is her own and that appropriate credit has been given where reference has been made to the work of others. This copy has been supplied on the understanding that it is copyright material and that no quotation from the thesis may be published without proper acknowledgement. The right of Clare Louise Wishart to be identified as Author of this work has been asserted by her in accordance with the Copyright, Designs and Patents Act 1988. © 2013 The University of Leeds and Clare Louise Wishart. II Acknowledgments Firstly I would like to offer my sincerest thanks and gratitude to my supervisor, Dr. Dan Donnelly, who has been nothing but encouraging and engaging from day one. I have thoroughly enjoyed every moment of working alongside him and learning from his guidance and wisdom. My thanks go to my academic assessor Professor Paul Milner whom I have known for several years, and during my time at the University of Leeds he has offered me invaluable advice and inspiration. Additionally I would like to thank my academic project advisor Dr. Michael Harrison for his friendship, help and advice. I would like to thank Dr. Rosalind Mann and Dr. Elsayed Nasr for welcoming me into the lab as a new PhD student and sharing their experimental techniques with me, these techniques have helped me no end in my time as a research student. -

P2Y6 Receptors Regulate CXCL10 Expression and Secretion in Mouse Intestinal Epithelial Cells
fphar-09-00149 February 26, 2018 Time: 17:57 # 1 ORIGINAL RESEARCH published: 28 February 2018 doi: 10.3389/fphar.2018.00149 P2Y6 Receptors Regulate CXCL10 Expression and Secretion in Mouse Intestinal Epithelial Cells Mabrouka Salem1,2, Alain Tremblay2, Julie Pelletier2, Bernard Robaye3 and Jean Sévigny1,2* 1 Département de Microbiologie-Infectiologie et d’Immunologie, Faculté de Médecine, Université Laval, Québec City, QC, Canada, 2 Centre de Recherche du CHU de Québec – Université Laval, Québec City, QC, Canada, 3 Institut de Recherche Interdisciplinaire en Biologie Humaine et Moléculaire, Université Libre de Bruxelles, Gosselies, Belgium In this study, we investigated the role of extracellular nucleotides in chemokine (KC, MIP- 2, MCP-1, and CXCL10) expression and secretion by murine primary intestinal epithelial cells (IECs) with a focus on P2Y6 receptors. qRT-PCR experiments showed that P2Y6 was the dominant nucleotide receptor expressed in mouse IEC. In addition, the P2Y6 Edited by: ligand UDP induced expression and secretion of CXCL10. For the other studies, we Kenneth A. Jacobson, −=− National Institutes of Health (NIH), took advantage of mice deficient in P2Y6 (P2ry6 ). Similar expression levels of P2Y1, −=− United States P2Y2, P2X2, P2X4, and A2A were detected in P2ry6 and WT IEC. Agonists of Reviewed by: TLR3 (poly(I:C)), TLR4 (LPS), P2Y1, and P2Y2 increased the expression and secretion Fernando Ochoa-Cortes, of CXCL10 more prominently in P2ry6−=− IEC than in WT IEC. CXCL10 expression Universidad Autónoma de San Luis −=− Potosí, Mexico and secretion induced by poly(I:C) in both P2ry6 and WT IEC were inhibited by Markus Neurath, general P2 antagonists (suramin and Reactive-Blue-2), by apyrase, and by specific Universitätsklinikum Erlangen, Germany antagonists of P2Y1, P2Y2, P2Y6 (only in WT), and P2X4. -

Immunosuppression Via Adenosine Receptor Activation by Adenosine
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Crossref RESEARCH ARTICLE elifesciences.org Immunosuppression via adenosine receptor activation by adenosine monophosphate released from apoptotic cells Hiroshi Yamaguchi1, Toshihiko Maruyama2, Yoshihiro Urade2†, Shigekazu Nagata1,3* 1Department of Medical Chemistry, Graduate School of Medicine, Kyoto University, Kyoto, Japan; 2Department of Molecular Behavioral Biology, Osaka Bioscience Institute, Osaka, Japan; 3Core Research for Evolutional Science and Technology, Japan Science and Technology Corporation, Kyoto, Japan Abstract Apoptosis is coupled with recruitment of macrophages for engulfment of dead cells, and with compensatory proliferation of neighboring cells. Yet, this death process is silent, and it does not cause inflammation. The molecular mechanisms underlying anti-inflammatory nature of the apoptotic process remains poorly understood. In this study, we found that the culture supernatant of apoptotic cells activated the macrophages to express anti-inflammatory genes such as Nr4a and Thbs1. A high level of AMP accumulated in the apoptotic cell supernatant in a Pannexin1-dependent manner. A nucleotidase inhibitor and A2a adenosine receptor antagonist inhibited the apoptotic *For correspondence: snagata@ supernatant-induced gene expression, suggesting AMP was metabolized to adenosine by an mfour.med.kyoto-u.ac.jp ecto-5’-nucleotidase expressed on macrophages, to activate the macrophage A2a adenosine receptor. Intraperitoneal injection of zymosan into Adora2a- or Panx1-deficient mice produced † Present address: Molecular high, sustained levels of inflammatory mediators in the peritoneal lavage. These results indicated Sleep Biology Laboratory, that AMP from apoptotic cells suppresses inflammation as a ‘calm down’ signal. WPI-International Institute for DOI: 10.7554/eLife.02172.001 Integrative Sleep Medicine, University of Tsukuba, Ibaraki, Japan Competing interests: The Introduction authors declare that no competing interests exist. -

Endothelial-Protective Effects of a G-Protein-Biased Sphingosine-1 Phosphate Receptor-1 Agonist, SAR247799, in Type-2 Diabetes R
medRxiv preprint doi: https://doi.org/10.1101/2020.05.15.20103101; this version posted May 20, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. All rights reserved. No reuse allowed without permission. 1 Endothelial-protective effects of a G-protein-biased sphingosine-1 phosphate receptor-1 agonist, SAR247799, in type-2 diabetes rats and a randomized placebo-controlled patient trial. Luc Bergougnan1, Grit Andersen2, Leona Plum-Mörschel3, Maria Francesca Evaristi1, Bruno Poirier1, Agnes Tardat4, Marcel Ermer3, Theresa Herbrand2, Jorge Arrubla2, Hans Veit Coester2, Roberto Sansone5, Christian Heiss6, Olivier Vitse4, Fabrice Hurbin4, Rania Boiron1, Xavier Benain4, David Radzik1, Philip Janiak1, Anthony J Muslin7, Lionel Hovsepian1, Stephane Kirkesseli1, Paul Deutsch8, Ashfaq A Parkar8 1 Sanofi R&D, 1 Avenue Pierre Brossolette, 91385 Chilly Mazarin, France; 2 Profil Institut für Stoffwechselforschung GmbH, Hellersbergstraße 9, 41460 Neuss, Germany; 3 Profil Mainz GmbH & Co. KG, Malakoff-Passage, Rheinstraße 4C, Eingang via Templerstraße, D-55116 Mainz, Germany; 4 Sanofi R&D, 371 Rue du Professeur Blayac, 34080 Montpellier, France; 5 University Hospital Düsseldorf, Division of Cardiology, Pulmonary diseases and Vascular medicine, 40225 Düsseldorf, Germany; 6 Department of Clinical and Experimental Medicine, University of Surrey, Stag Hill, Guildford GU2 7XH, UK; 7 Sanofi US Services, 640 Memorial Drive, Cambridge MA 02139, USA; 8 Sanofi US Services, 55 Corporate Drive, Bridgewater, NJ 08807, USA. The authors confirm that the Principal Investigators for the clinical study were Grit Anderson and Leona Plum- Mörschel and that they had direct clinical responsibility for patients at the Neuss and Mainz sites, respectively. -
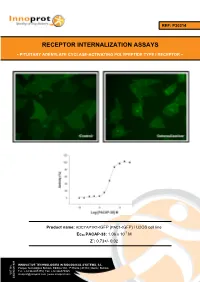
Receptor Internalization Assays
REF: P30214 RECEPTOR INTERNALIZATION ASSAYS - PITUITARY ADENYLATE CYCLASE-ACTIVATING POLYPEPTIDE TYPE I RECEPTOR - Product name: ADCYAP1R1-tGFP (PAC1-tGFP) / U2OS cell line -7 Ec50 PACAP-38: 1.06 x 10 M Z´: 0.73+/- 0.02 INNOVATIVE TECHNOLOGIES IN BIOLOGICAL SYSTEMS, S.L. Parque Tecnológico Bizkaia, Edifício 502, 1ª Planta | 48160 | Derio | Bizkaia Tel.: +34 944005355 | Fax: +34 946579925 VAT No. [email protected] | www.innoprot.com ESB95481909 Product Name: ADCYAP1R1-tGFP_U2OS Reference: P30214 Rep. Official Full Name: Pituitary adenylate cyclase- activating polypeptide type I receptor DNA Accession Number: Gene Bank AY366498 Host Cell: U2OS References: P30214: 2 vials of 3 x 106 proliferative cells P30214-DA: 1 vial of 2 x 106 division-arrested cells Storage: Liquid Nitrogen Assay Briefly description About ADCYAP1R1 Each vial of ADCYAP1R1 Internalization Assay Pituitary adenylate cyclase-activating Cell Line contains U2OS cells stably expressing polypeptide type I receptor, also known as human Pituitary adenylate cyclase-activating PAC1 is a protein that in humans is encoded by polypeptide type I receptor tagged in the N- the ADCYAP1R1 gene. ADCYAP1R1 is a terminus with tGFP protein. membrane-associated protein and shares significant homology with members of the Innoprot’s ADCYAP1R1-tGFP Internalization glucagon/secretin receptor family. This receptor Assay Cell Line has been designed to assay binds pituitary adenylate cyclase activating potential agonists/ antagonists against peptide (PACAP) mediating several biological ADCYAP1R1, modulating its activation and the activities and it is positively coupled to following redistribution process inside the cells. adenylate cyclase. This cell line will allow the image analysis of the stimuli induced by the compounds. -

Molecular Dissection of G-Protein Coupled Receptor Signaling and Oligomerization
MOLECULAR DISSECTION OF G-PROTEIN COUPLED RECEPTOR SIGNALING AND OLIGOMERIZATION BY MICHAEL RIZZO A Dissertation Submitted to the Graduate Faculty of WAKE FOREST UNIVERSITY GRADUATE SCHOOL OF ARTS AND SCIENCES in Partial Fulfillment of the Requirements for the Degree of DOCTOR OF PHILOSOPHY Biology December, 2019 Winston-Salem, North Carolina Approved By: Erik C. Johnson, Ph.D. Advisor Wayne E. Pratt, Ph.D. Chair Pat C. Lord, Ph.D. Gloria K. Muday, Ph.D. Ke Zhang, Ph.D. ACKNOWLEDGEMENTS I would first like to thank my advisor, Dr. Erik Johnson, for his support, expertise, and leadership during my time in his lab. Without him, the work herein would not be possible. I would also like to thank the members of my committee, Dr. Gloria Muday, Dr. Ke Zhang, Dr. Wayne Pratt, and Dr. Pat Lord, for their guidance and advice that helped improve the quality of the research presented here. I would also like to thank members of the Johnson lab, both past and present, for being valuable colleagues and friends. I would especially like to thank Dr. Jason Braco, Dr. Jon Fisher, Dr. Jake Saunders, and Becky Perry, all of whom spent a great deal of time offering me advice, proofreading grants and manuscripts, and overall supporting me through the ups and downs of the research process. Finally, I would like to thank my family, both for instilling in me a passion for knowledge and education, and for their continued support. In particular, I would like to thank my wife Emerald – I am forever indebted to you for your support throughout this process, and I will never forget the sacrifices you made to help me get to where I am today. -

Supplementary Table 3 Complete List of RNA-Sequencing Analysis of Gene Expression Changed by ≥ Tenfold Between Xenograft and Cells Cultured in 10%O2
Supplementary Table 3 Complete list of RNA-Sequencing analysis of gene expression changed by ≥ tenfold between xenograft and cells cultured in 10%O2 Expr Log2 Ratio Symbol Entrez Gene Name (culture/xenograft) -7.182 PGM5 phosphoglucomutase 5 -6.883 GPBAR1 G protein-coupled bile acid receptor 1 -6.683 CPVL carboxypeptidase, vitellogenic like -6.398 MTMR9LP myotubularin related protein 9-like, pseudogene -6.131 SCN7A sodium voltage-gated channel alpha subunit 7 -6.115 POPDC2 popeye domain containing 2 -6.014 LGI1 leucine rich glioma inactivated 1 -5.86 SCN1A sodium voltage-gated channel alpha subunit 1 -5.713 C6 complement C6 -5.365 ANGPTL1 angiopoietin like 1 -5.327 TNN tenascin N -5.228 DHRS2 dehydrogenase/reductase 2 leucine rich repeat and fibronectin type III domain -5.115 LRFN2 containing 2 -5.076 FOXO6 forkhead box O6 -5.035 ETNPPL ethanolamine-phosphate phospho-lyase -4.993 MYO15A myosin XVA -4.972 IGF1 insulin like growth factor 1 -4.956 DLG2 discs large MAGUK scaffold protein 2 -4.86 SCML4 sex comb on midleg like 4 (Drosophila) Src homology 2 domain containing transforming -4.816 SHD protein D -4.764 PLP1 proteolipid protein 1 -4.764 TSPAN32 tetraspanin 32 -4.713 N4BP3 NEDD4 binding protein 3 -4.705 MYOC myocilin -4.646 CLEC3B C-type lectin domain family 3 member B -4.646 C7 complement C7 -4.62 TGM2 transglutaminase 2 -4.562 COL9A1 collagen type IX alpha 1 chain -4.55 SOSTDC1 sclerostin domain containing 1 -4.55 OGN osteoglycin -4.505 DAPL1 death associated protein like 1 -4.491 C10orf105 chromosome 10 open reading frame 105 -4.491 -

Bradykinin Receptor Deficiency Or Antagonism Do Not Impact the Host
Ding et al. Intensive Care Medicine Experimental (2019) 7:14 Intensive Care Medicine https://doi.org/10.1186/s40635-019-0228-3 Experimental RESEARCH Open Access Bradykinin receptor deficiency or antagonism do not impact the host response during gram-negative pneumonia-derived sepsis Chao Ding1,2, Jack Yang2, Cornelis van’t Veer2 and Tom van der Poll2,3* * Correspondence: t.vanderpoll@ amc.uva.nl Abstract 2Center of Experimental and Molecular Medicine, Academic Background: Kinins are short peptides with a wide range of proinflammatory Medical Center, University of properties that are generated from kininogens in the so-called kallikrein-kinin system. Amsterdam, Meibergdreef 9, Room Kinins exert their biological activities through stimulation of two distinct receptor G2-130, 1105 AZ Amsterdam, the Netherlands subtypes, the kinin or bradykinin B1 and B2 receptors (B1R, B2R). Acute challenge 3Division of Infectious Diseases, models have implicated B1R and B2R in the pathogenesis of sepsis. However, their Academic Medical Center, role in the host response during sepsis originating from the lung is not known. University of Amsterdam, Amsterdam, the Netherlands Results: To determine the role of B1R and B2R in pneumonia-derived sepsis, B1R/ Full list of author information is B2R-deficient mice and wild-type mice treated with the B1R antagonist R-715 or the available at the end of the article B2R antagonist HOE-140 were studied after infection with the common gram- negative pathogen Klebsiella pneumoniae via the airways. Neither B1R/B2R deficiency nor B1R or B2R inhibition influenced bacterial growth at the primary site of infection or dissemination to distant body sites. -

An Explanation of G-Protein Coupled Receptor Structure
Journal of Stem Cell Research & Therapeutics Review Article Open Access Role of GPCRS towards cell: an explanation of g-protein coupled receptor structure Abstract Volume 2 Issue 3 - 2017 G- protein coupled receptors are the heptahelical, serpentine receptor and are Nidhi Sharma,1 Anil Kumar Sahdev,2 Vinit Raj2 multifunctional receptors having modulatory activity of a wide variety of biological 1Dr. K. N. Modi Institute of Pharmacy & Research centre, process including: Neurotransmission, Chemoattraction, Cardiac function, Olfaction Modinagar, India. and Vision etc. At largest level they are involved in signal transduction across cell 2Department of Pharmaceutical Sciences, Babasaheb Bhimrao membranes therefore they represent major targets in the development of novel drug Ambedkar University, India candidates in all clinical areas. Particularly membrane cholesterol has been reported to have a great role in the functioning of a number of GPCRs. Apart from this it also Correspondence: Vinit Raj, Department of Pharmaceutical have some drawbacks. Mutations that occur are associated with a broad spectrum of Sciences, Babasaheb Bhimrao Ambedkar University, VidyaVihar, diseases of diverse etiology. As a mutations result, there is a change in receptor activity Rae Bareli Road, Lucknow-226025, Email [email protected] (GPCR become inactive, overactive, or constitutively active), in the process of ligand binding and signal transduction. Changes in the GPCRs functioning can cause diseases Received: January 31, 2017 | Published: March 27, 2017 such as retinitis pigmentosa (rhodopsin mutations), nephrogenic diabetes insipidus (vasopressin receptor mutations), and obesity (melanocortin receptor mutations). Keywords: gpcrs, serpentine receptor, neurotransmission, retinitis pigmentosa, nephrogenic diabetes insipidus, bardet-biedl syndrome, hypogonadism Introduction deliver a message and appropriate cellular and physiological response1,2 (Figure 1). -

A2B Adenosine Receptors and T Cell Activation 493
Journal of Cell Science 112, 491-502 (1999) 491 Printed in Great Britain © The Company of Biologists Limited 1999 JCS0069 Expression of A2B adenosine receptors in human lymphocytes: their role in T cell activation Maribel Mirabet1, Carolina Herrera1, Oscar J. Cordero2, Josefa Mallol1, Carmen Lluis1 and Rafael Franco1,* 1Department of Biochemistry and Molecular Biology, Faculty of Chemistry, University of Barcelona, Barcelona, Catalonia, Spain 2Department of Biochemistry and Molecular Biology, Faculty of Biology, University of Santiago de Compostela, Spain *Author for correspondence (e-mail: [email protected]; homepage: www.bq.ub.es/recep/franco.html) Accepted 9 December 1998; published on WWW 25 January 1999 SUMMARY Extracellular adenosine has a key role in the development A2BRs but not of A2A receptors in these human cells. The and function of the cells of the immune system. Many of percentage of A2BR-expressing cells was similar in the the adenosine actions seem to be mediated by specific CD4+ or CD8+ T cell subpopulations. Interestingly surface receptors positively coupled to adenylate cyclase: activation signals delivered by either phytohemagglutinin A2A and A2B. Despite the fact that A2A receptors (A2ARs) or anti-T cell receptor/CD3 complex antibodies led to a can be easily studied due to the availability of the specific significant increase in both the percentage of cells agonist CGS21680, a pharmacological and physiological expressing the receptor and the intensity of the labeling. characterization of adenosine A2B receptors (A2BRs) in These receptors are functional since interleukin-2 lymphocytes has not been possible due to the lack of production in these cells is reduced by NECA but not by R- suitable reagents. -

Binding Mode Exploration of B1 Receptor Antagonists' by the Use of Molecular Dynamics and Docking Simulation—How Different T
International Journal of Molecular Sciences Article Binding Mode Exploration of B1 Receptor Antagonists’ by the Use of Molecular Dynamics and Docking Simulation—How Different Target Engagement Can Determine Different Biological Effects Marica Gemei 1,*, Carmine Talarico 1 , Laura Brandolini 1, Candida Manelfi 1, Lorena Za 2, Silvia Bovolenta 2, Chiara Liberati 2, Luigi Del Vecchio 3, Roberto Russo 4 , Carmen Cerchia 4, Marcello Allegretti 1 and Andrea Rosario Beccari 1 1 Dompé Farmaceutici SpA, via Campo di Pile, 67100 L’Aquila, Italy; [email protected] (C.T.); [email protected] (L.B.); candida.manelfi@dompe.com (C.M.); [email protected] (M.A.); [email protected] (A.R.B.) 2 Axxam, Via Meucci 3, Bresso, 20091 Milano, Italy; [email protected] (L.Z.); [email protected] (S.B.); [email protected] (C.L.) 3 Ceinge Biotecnologie Avanzate, via G. Salvatore 486, 80145 Napoli, Italy; [email protected] 4 Department of Pharmacy, University of Naples “Federico II”, via D. Montesano, 49, 80131 Napoli, Italy; [email protected] (R.R.); [email protected] (C.C.) * Correspondence: [email protected]; Tel.: +34-06-465916 Received: 26 August 2020; Accepted: 12 October 2020; Published: 16 October 2020 Abstract: The kinin B1 receptor plays a critical role in the chronic phase of pain and inflammation. The development of B1 antagonists peaked in recent years but almost all promising molecules failed in clinical trials. Little is known about these molecules’ mechanisms of action and additional information will be necessary to exploit the potential of the B1 receptor. -

Adenosine Receptors and Cancer
Adenosine Receptors and Cancer P. Fishman, S. Bar-Yehuda, M. Synowitz, J.D. Powell, K.N. Klotz, S. Gessi, and P.A. Borea Contents 1 Introduction..................................................................................... 402 2A1 Adenosine Receptor........................................................................ 402 3A2A Adenosine Receptor...................................................................... 406 3.1 The A2AAR:ProtectorofHostTissue,ProtectorofTumors......................... 406 3.2 TumorsEvadetheImmuneSystembyInhibitingImmuneCellFunction........... 406 3.3 The A2AAR Negatively Regulates Immune Responses............................... 407 3.4 AdenosineProtectsTumorsfromImmuneDestruction............................... 408 3.5 A2AAR Antagonism as a Means of Enhancing Immunotherapy..................... 410 4A2B Adenosine Receptors..................................................................... 410 5A3 Adenosine Receptor........................................................................ 414 5.1 Overexpression of the A3AR in Tumor Versus Normal Adjacent Tissues........... 415 5.2 InVitroStudies......................................................................... 417 5.3 InVivoStudies.......................................................................... 419 5.4 Mechanisms of Action for the Anticancer Activity of the A3AR.................... 424 6 Anticancer Activity of A3AR Antagonists.................................................... 429 7 SummaryandConclusions...................................................................