Isolation and Identification of Pathogenic Bacteria in Edible Fish
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Research and Investigations in Chilika Lake (1872 - 2017)
Bibliography of Publications Research and Investigations in Chilika Lake (1872 - 2017) Surya K. Mohanty Krupasindhu Bhatta Susanta Nanda 2018 Chilika Development Authority Chilika Development Authority Forest & Environment Department, Government of Odisha, Bhubaneswar Bibliography of Publications Research and Investigations in Chilika Lake (1872 - 2017) Copyright: © 2018 Chilika Development Authority, C-11, B.J.B. Nagar, Bhubaneswar - 751 014 Copies available from: Chilika Development Authority (A Government of Odisha Agency) C-11, B.J.B. Nagar Bhubaneswar - 751 014 Tel: +91 674 2434044 / 2436654 Fax: +91 674 2434485 Citation: Mohanty, Surya K., Krupasindhu Bhatta and Susanta Nanda (2018). Bibliography of Publications: Research and Investigations in Chilika Lake (1872–2017). Chilika Development Authority, Bhubaneswar : 190 p. Published by: Chief Executive, Chilika Development Authority, C-11, B.J.B. Nagar, Bhubaneswar - 751 014 Design & Print Third Eye Communications Bhubaneswar [email protected] Foreword Chilika Lake with unique ecological character featured by amazing biodiversity and rich fishery resources is the largest brackishwater lake in Asia and the second largest in the world. Chilika with its unique biodiversity wealth, ecological diversity and being known as an avian paradise is the pride of our wetland heritage and the first designated Indian Ramsar Site. The ecosystem services of Chilika are critical to the functioning of our life support system in general and livelihood of more than 0.2 million local fishers and other stakeholders in particular. It is also one of the few lakes in the world which sustain the population of threatened Irrawaddy Dolphin. Chilika also has a long history of its floral and faunal studies which begun since more than a century ago. -
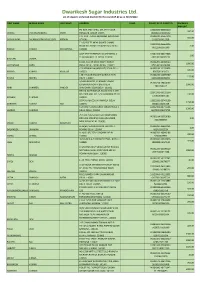
Dwarikesh Sugar Industries Ltd. List of Unpaid / Unclaimed Dividend for the Year 2019-20 As on 31/03/2020
Dwarikesh Sugar Industries Ltd. List of unpaid / unclaimed dividend for the year 2019-20 as on 31/03/2020 FIRST NAME MIDDLE NAME LAST NAME ADDRESS FOLIO FOLIO/ DP ID CLIENT ID DIVIDEND AMOUNT PO BOX 3747 SHELL GTL 9TH FLOOR 12032800-00485214- 300.00 CHIRAG PRAVINCHANDRA SHAH MIRQUAB TOWER DOHA 0602321270000152 P.O. BOX - 13027 ABUDHABI ABUDHABI IN300239-14567240- 550.00 GOPAKUMAR PALAMADATHUVASUDEVA MENON OTHERS 10132100012128 T-166 FIST FF NEAR GULATI CHAKKI 12029900-05661905- ROAD NO 20 BALJIT NAGAR New Delhi 2.00 00111000161340 PANKAJ KUMAR SRIVASTAVA 110008 A/67 RESETTLEMENT COLONY KHYALA 12082500-00422908- 1.00 TILAK NAGAR S. O DELHI 110018 603100100005239 BHUVAN VOHRA B-325, SOUTH MOTI BAGH, NANAK IN302269-11080027- 1000.00 CHITRANSHI SAND PURA, DELHI, DELHI, INDIA. 110021 073100100326388 C 20 KIDWAI NAGAR(EAST) DELHI DELHI IN300239-12178458- 500.00 ARVIND KUMAR KHULLAR 110023 9007201006173 J 36 II FLOOR RAJOURI GARDEN NEW IN302236-11096760- 125.00 PAVAN MEHRA DELHI 110027 040104000054269 1/5340 GALI NO 14 BALBIR NAGAR IN300214-19464000- EXTENTION NORTH EAST DELHI 1000.00 7611712117 HARI SHANKER PANDEY SHAHDARA DELHI DELHI 110032 BHOLA NATH NAGAR RAMA BLOCK QTR 12041900-00129504- NO-1900 GALI NO. 5, SHAHADARA DELHI 100.00 13501000005787 NEELAM SHARMA 110032 56 PUNJABI COLONY NARELA DELHI 12033200-05743199- 1250.00 DARSHAN KUMAR HUF 110040 00201110105208 C 29 IIND FLOOR ASHOK VIHAR PHASE 1 12032300-00742101- 1300.00 SHARAD SHARMA DELHI DELHI 110052 0637000104271791 79 C LIG FLATS DDA FLAT MADHUBAN IN300214-20053080- ENCLAVE MADIPUR -

Banco 2016-17 Final
Note: This sheet is applicable for uploading the particulars related to the unclaimed and unpaid amount pending with company. Make sure that the details are in accordance with the information already provided in e-form IEPF-2 Date Of AGM(DD-MON-YYYY) CIN/BCIN L51100GJ1961PLC001039 Prefill Company/Bank Name BANCO PRODUCTS (INDIA) LIMITED 22-Sep-2018 Sum of unpaid and unclaimed dividend 1930984.00 Sum of interest on matured debentures 0.00 Sum of matured deposit 0.00 Sum of interest on matured deposit 0.00 Sum of matured debentures 0.00 Sum of interest on application money due for refund 0.00 Sum of application money due for refund 0.00 Redemption amount of preference shares 0.00 Sales proceed for fractional shares 0.00 Validate Clear Proposed Date of Investor First Investor Middle Investor Last Father/Husband Father/Husband Father/Husband Last DP Id-Client Id- Amount Address Country State District Pin Code Folio Number Investment Type transfer to IEPF Name Name Name First Name Middle Name Name Account Number transferred (DD-MON-YYYY) PUNAMBHAI B PATEL NA NA NA SHRI LAXMI BADAMI COAL FACTORY,INDIA INDUSTRY'S COMPOUND, GUJARAT PRATAPNAGAR, VADODARA. VADODARA 39000400000026 Amount for unclaimed and unpaid dividend1200.00 22-NOV-2024 A NA JAGANNATHANNA NA NA NO-11, BAKTHAWATCHALAM NAGAR,INDIA III STREET, MADRAS CHENNAI TAMIL NADU CHENNAI 60002000010003 Amount for unclaimed and unpaid dividend400.00 22-NOV-2024 ABDUL NA RASHID NA NA NA I/II MAKER TOWERS, CUFFE PARADE,INDIA MUMBAI. MUMBAI MAHARASHTRA MUMBAI 40000500010018 Amount for unclaimed and unpaid dividend1000.00 22-NOV-2024 AMRITLAL NA MONGA NA NA NA 31,PRATAP COLONY, MODEL GRAM, INDIALUDHIANA. -

Problems of Salination of Land in Coastal Areas of India and Suitable Protection Measures
Government of India Ministry of Water Resources, River Development & Ganga Rejuvenation A report on Problems of Salination of Land in Coastal Areas of India and Suitable Protection Measures Hydrological Studies Organization Central Water Commission New Delhi July, 2017 'qffif ~ "1~~ cg'il'( ~ \jf"(>f 3mft1T Narendra Kumar \jf"(>f -«mur~' ;:rcft fctq;m 3tR 1'j1n WefOT q?II cl<l 3re2iM q;a:m ~0 315 ('G),~ '1cA ~ ~ tf~q, 1{ffit tf'(Chl '( 3TR. cfi. ~. ~ ~-110066 Chairman Government of India Central Water Commission & Ex-Officio Secretary to the Govt. of India Ministry of Water Resources, River Development and Ganga Rejuvenation Room No. 315 (S), Sewa Bhawan R. K. Puram, New Delhi-110066 FOREWORD Salinity is a significant challenge and poses risks to sustainable development of Coastal regions of India. If left unmanaged, salinity has serious implications for water quality, biodiversity, agricultural productivity, supply of water for critical human needs and industry and the longevity of infrastructure. The Coastal Salinity has become a persistent problem due to ingress of the sea water inland. This is the most significant environmental and economical challenge and needs immediate attention. The coastal areas are more susceptible as these are pockets of development in the country. Most of the trade happens in the coastal areas which lead to extensive migration in the coastal areas. This led to the depletion of the coastal fresh water resources. Digging more and more deeper wells has led to the ingress of sea water into the fresh water aquifers turning them saline. The rainfall patterns, water resources, geology/hydro-geology vary from region to region along the coastal belt. -

Kerala State Pollution Control Board
: General: 0471- 2312910, 2318153, 2318154, 2318155 Chairman: 2318150 Member Secretary: 2318151 e-mail: [email protected] FAX: 0471 – 2318134, 2318152 web: www.keralapcb.nic.in KERALA STATE POLLUTION CONTROL BOARD Pattom P.O., Thiruvananthapuram – 695 004 PCB/HO/EE3/NGT/673/2018 Date:4.08.2020 From The Member Secretary Kerala State Pollution Control Board To The Secretary Ministry of Jal Shakti Department of water Resources, River Development and Ganga Rejuvenation Shram Shakti Bhawan Rafi Marg New Delhi-110001 Subject: Forwarding of Monthly Progress Report of June 2020 – reg. Ref: Copy of presentation made on 30.07.2020 in the 4th meeting of CMC held through Video Conferencing regarding 351 polluted river stretches based on the directions of Hon’ble NGT in the matter OA No. 673 of 2018, received through e-mail on 31.07.2020 Sir, The Monthly Progress Reports of June 2020 regarding all the 21 polluted river stretches in Kerala based on the directions of Hon’ble NGT in the matter OA No. 673 of 2018 were submitted through e-mail on 18.07.2020 to [email protected] , [email protected], [email protected], [email protected], [email protected] and [email protected]. Now in compliance with the instructions forwarded along with the copy of presentation, which was made on 30.07.2020 in the 4th meeting of CMC held under the Chairmanship of Secretary, DoWR,RD&GR, Ministry of Jal Shakti, received through e- mail on 31.07.2020, as “MPRs are submitted in 40 different files in word and excel format and State to submit one signed compiled PDF copy of MPR to NMCG,” signed compiled PDF copy of MPR is submitted herewith for kind information and necessary action. -

Etroplus Suratensis) Ecological Risk Screening Summary
Green Chromide Cichlid (Etroplus suratensis) Ecological Risk Screening Summary U.S. Fish and Wildlife Service, April 2011 Revised, September 2018 Web Version, 6/5/2019 Photo: P. Corbett. Licensed under CC BY 2.0. Available: https://flic.kr/p/tmiiei. (September 2018). 1 Native Range and Status in the United States Native Range From Froese and Pauly (2018): “Western Indian Ocean: India and Sri Lanka.” 1 From Abraham (2011): “Etroplus suratensis is distributed in the coastal regions of peninsular India and Sri Lanka. In India, the wild populations have been recorded from the states of Kerala and Tamil Nadu.” Status in the United States This species has not been reported as introduced or established in the United States. This species is in trade in the United States. From Imperial Tropicals (2015): “Green Chromide Cichlid (Etroplus suratensis) […] $ 19.99” From Bluegrass Aquatics (2019): “Green Chromide Cichlid – REGULAR $26.98” Means of Introductions in the United States This species has not been reported as introduced or established in the United States. 2 Biology and Ecology Taxonomic Hierarchy and Taxonomic Standing From ITIS (2018): “Kingdom Animalia Subkingdom Bilateria Infrakingdom Deuterostomia Phylum Chordata Subphylum Vertebrata Infraphylum Gnathostomata Superclass Actinopterygii Class Teleostei Superorder Acanthopterygii Order Perciformes Suborder Labroidei Family Cichlidae Genus Etroplus Species Etroplus suratensis (Bloch, 1790)” From Fricke et al. (2018): “Current status: Valid as Etroplus suratensis (Bloch 1790). Cichlidae: Etroplinae.” 2 Size, Weight, and Age Range From Froese and Pauly (2018): “Max length : 40.0 cm TL male/unsexed; [Menon 1999]; common length : 20.0 cm TL male/unsexed; [Pethiyagoda 1991]” Environment From Froese and Pauly (2018): “Brackish; benthopelagic; depth range 10 - ? m. -
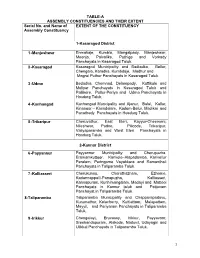
List of Lacs with Local Body Segments (PDF
TABLE-A ASSEMBLY CONSTITUENCIES AND THEIR EXTENT Serial No. and Name of EXTENT OF THE CONSTITUENCY Assembly Constituency 1-Kasaragod District 1 -Manjeshwar Enmakaje, Kumbla, Mangalpady, Manjeshwar, Meenja, Paivalike, Puthige and Vorkady Panchayats in Kasaragod Taluk. 2 -Kasaragod Kasaragod Municipality and Badiadka, Bellur, Chengala, Karadka, Kumbdaje, Madhur and Mogral Puthur Panchayats in Kasaragod Taluk. 3 -Udma Bedadka, Chemnad, Delampady, Kuttikole and Muliyar Panchayats in Kasaragod Taluk and Pallikere, Pullur-Periya and Udma Panchayats in Hosdurg Taluk. 4 -Kanhangad Kanhangad Muncipality and Ajanur, Balal, Kallar, Kinanoor – Karindalam, Kodom-Belur, Madikai and Panathady Panchayats in Hosdurg Taluk. 5 -Trikaripur Cheruvathur, East Eleri, Kayyur-Cheemeni, Nileshwar, Padne, Pilicode, Trikaripur, Valiyaparamba and West Eleri Panchayats in Hosdurg Taluk. 2-Kannur District 6 -Payyannur Payyannur Municipality and Cherupuzha, Eramamkuttoor, Kankole–Alapadamba, Karivellur Peralam, Peringome Vayakkara and Ramanthali Panchayats in Taliparamba Taluk. 7 -Kalliasseri Cherukunnu, Cheruthazham, Ezhome, Kadannappalli-Panapuzha, Kalliasseri, Kannapuram, Kunhimangalam, Madayi and Mattool Panchayats in Kannur taluk and Pattuvam Panchayat in Taliparamba Taluk. 8-Taliparamba Taliparamba Municipality and Chapparapadavu, Kurumathur, Kolacherry, Kuttiattoor, Malapattam, Mayyil, and Pariyaram Panchayats in Taliparamba Taluk. 9 -Irikkur Chengalayi, Eruvassy, Irikkur, Payyavoor, Sreekandapuram, Alakode, Naduvil, Udayagiri and Ulikkal Panchayats in Taliparamba -

Indian Red Cross Society, D.K District Branch Life Members Details As on 02.10.2015
Indian Red Cross Society, D.K District Branch Life Members details as on 02.10.2015 Sri. J.R. Lobo, Sri. RTN. P.H.F William M.L.A, D'Souza, Globe Travels, Deputy Commissioner Jency, Near Ramakrishna 1 2 3 G06, Souza Arcade, Balmatta D.K District Tennis Court, 1st cross, Shiva Road, Mangalore-2 Bagh, Kadri, M’lore – 2 Ph: 9845080597 Ph: 9448375245 Sri. RTN. Nithin Shetty, Rtn. Sathish Pai B. Rtn. Ramdas Pai, 301, Diana APTS, S.C.S 4 5 Bharath Carriers, N.G Road 6 Pais Gen Agencies Port Road, Hospital Road, Balmatta, Attavar, Mangalore - 1 Bunder, Mangalore -1 Mangalore - 2 Sri. Vijaya Kumar K, Rtn. Ganesh Nayak, Rtn. S.M Nayak, "Srishti", Kadri Kaibattalu, Nayak & Pai Associates, C-3 Dukes Manor Apts., 7 8 9 D.No. 3-19-1691/14, Ward Ganesh Kripa Building, Matadakani Road, No. 3 (E), Kadri, Mangalore Carstreet, Mangalore 575001 Urva, Mangalore- 575006 9844042837 Rtn. Narasimha Prabhu RTN. Ashwin Nayak Sujir RTN. Padmanabha N. Sujir Vijaya Auto Stores "Varamahalaxmi" 10 "Sri Ganesh", Sturrock Road, 11 12 New Ganesh Mahal, 4-5-496, Karangalpady Cross Falnir, Mangalore - 575001 Alake, Mangalore -3 Road, Mangalore - 03 RTN. Rajendra Shenoy Rtn. Arun Shetty RTN. Rajesh Kini 4-6-615, Shivam Block, Excel Engineers, 21, Minar 13 14 "Annapoorna", Britto Lane, 15 Cellar, Saimahal APTS, Complex New Balmatta Road, Falnir, Mangalore - 575001 Karangalpady, Mangalore - 03 Mangalore - 1 Sri. N.G MOHAN Ravindranath K RTN. P.L Upadhya C/o. Beta Agencies & Project 803, Hat Hill Palms, Behind "Sithara", Behind K.M.C Private Ltd., 15-12-676, Mel Indian Airlines, Hat Hill Bejai, 16 17 18 Hospital, Attavar, Nivas Compound, Kadri, Mangalore – 575004 Mangalore - 575001 Mangalore – 02. -

Indian and Madagascan Cichlids
FAMILY Cichlidae Bonaparte, 1835 - cichlids SUBFAMILY Etroplinae Kullander, 1998 - Indian and Madagascan cichlids [=Etroplinae H] GENUS Etroplus Cuvier, in Cuvier & Valenciennes, 1830 - cichlids [=Chaetolabrus, Microgaster] Species Etroplus canarensis Day, 1877 - Canara pearlspot Species Etroplus suratensis (Bloch, 1790) - green chromide [=caris, meleagris] GENUS Paretroplus Bleeker, 1868 - cichlids [=Lamena] Species Paretroplus dambabe Sparks, 2002 - dambabe cichlid Species Paretroplus damii Bleeker, 1868 - damba Species Paretroplus gymnopreopercularis Sparks, 2008 - Sparks' cichlid Species Paretroplus kieneri Arnoult, 1960 - kotsovato Species Paretroplus lamenabe Sparks, 2008 - big red cichlid Species Paretroplus loisellei Sparks & Schelly, 2011 - Loiselle's cichlid Species Paretroplus maculatus Kiener & Mauge, 1966 - damba mipentina Species Paretroplus maromandia Sparks & Reinthal, 1999 - maromandia cichlid Species Paretroplus menarambo Allgayer, 1996 - pinstripe damba Species Paretroplus nourissati (Allgayer, 1998) - lamena Species Paretroplus petiti Pellegrin, 1929 - kotso Species Paretroplus polyactis Bleeker, 1878 - Bleeker's paretroplus Species Paretroplus tsimoly Stiassny et al., 2001 - tsimoly cichlid GENUS Pseudetroplus Bleeker, in G, 1862 - cichlids Species Pseudetroplus maculatus (Bloch, 1795) - orange chromide [=coruchi] SUBFAMILY Ptychochrominae Sparks, 2004 - Malagasy cichlids [=Ptychochrominae S2002] GENUS Katria Stiassny & Sparks, 2006 - cichlids Species Katria katria (Reinthal & Stiassny, 1997) - Katria cichlid GENUS -

Victorian Cichlid Society Incorporated
tthehe Did you hear that somebody cichlid mmonthlyonthly really cool is going to advertise here? As cool as us? . Used with permission David Callele Picture Copyright 2007 Victorian Cichlid Society Incorporated 36:04, May 2007 $1.10 Certificate of Incorporation # A0012794D REGISTERED BY AUSTRALIA POST - PP342780/0024 do not accept that there is anyone 20 I who is actually incapable of CCOMMITTEE:OMMITTEE: 1 writing an article about their fi sh, or ccichlidichlidGGetet youryour TCMTCM TThehe PRESIDENT: experiences with fi sh. I do, however, John McCormick ......................5944 3502 know how much I hate writing [email protected] Editorials. They are usually left to last sscenecene ICE RESIDENT LLastast V -P : and often hurried. This method has Klaus Schwarzenholz .........0414 444 737 landed me in “trouble” in the past THE NEXT MEETING of the Society willLLiving iving CColourolour due to dotting a few “t”s, crossing be held on the first Wednesday of the SECRETARY: WWordord month at 8 pm sharp (the Trading Table Graham Rowe ...........................9560-7472 an “i” or two, and in several cases, [email protected] Daryl Hutchins.. misplacing a comma. opens earlier) in the Mitcham Scout Hall, ggoo ttoo Brunswick Road, Mitcham. Visitors are TREASURER: A much better method that I have heartily encouraged to come along.hhttp://home.vicnet.net.au/ttp://hTonyom Fergusone.v i....................0408cnet.ne 533t. 552au/ s a result of recent discussions found is to write small notes in my CICHLID OF THE MONTH: Trout Cichlid Aat meetings both General and committee diary (yes, I jot down all ~~cichlid/MagList.htcichEDITORlid: /MagList.htm Committee, and weighing all the pros - Tony Ferguson. -

1. Introduction
1. INTRODUCTION 1.1. History Kasaragod, the northernmost district of Kerala, is endowed with rich natural resources and is noted for its majestic forts, ravishing rivers, hills, green valleys and beautiful beaches. The rich and varied cultural heritage of the district is portrayed through spectacular presentations of Theyyam, Yakshagana, Poorakkali, Kolkali and Mappilappattu. Seven languages are prevalent in Kasaragod. Malayalam is the administrative language. Other languages are Kannada, Tulu, Konkani, Marati, Urdu and Beary.Prior to State reorganization, Kasaragod was part of the South Kanara district.Kasaragod became a part of Malabar district following the reorganization of States and formation of unified Kerala State. Later, Kasaragod Taluk of Malabar district was bifurcated in to Kasaragod and Hosdurg Taluks and integrated with the then newly formed Cannanore district. Kasaragod became part of Kerala following the re-organization of states and formation of Kerala in 1st November 1956.The district was Kasaragod Taluk in Kannur District.The formation of Kasaragod district was a long felt ambition of the people.It is with the intention of bestowing maximum attention on the development of backward area, Kasaragod district was formed on 24th May,1984 as per GO (MS)No.520/84/RD, Dated 19.05.1984 by taking Kasaragod and Hosdurg taluks from the then Kannur district.The name Kasaragod is said to be derived from the word Kasaragod which means Nuxvemied Forest(Kanjirakuttam). 1.2. Physiography Kasaragod is bounded on the north and the east by Dakshina Kannada and Coorg districts of Karnataka State, on the south by Kannur district and on the west by the Lakshadweep Sea. -

PROCEEDINGS of the DISTRICT COLLECTOR, KASARAGOD (Present: Dr
PROCEEDINGS OF THE DISTRICT COLLECTOR, KASARAGOD (Present: Dr. D. Sajith Babu, IAS.) Sub:- Kasaragod Development Package 2020-21- Integrated water management of Kasaragod district- Renovation of VCB at Edaneer across Mogral river in Chengala G.P - Administrative Sanction– Order issued- Reg. Read:- 1. G.O (Rt) No.392/201 3/Plg Dated: 10.10.2013 2. G.O (Rt) No. 337/2019/P&EA Dated: 25.07.2019 3. Letter No. D1-KDP/2020 Dated: 05.10.2020 of the Executive Engineer, Minor Irrigation Division, Kasaragod 4. Letter No. 431/2020/KDP/KSD Dated: 21.10.2020 of the District Collector, Kasaragod 5. Letter No. SPB/2312/2012/Agri Dated: 23.11.2020 of the Member Secretary, State Planning Board, Thiruvananthapuram 6. Minutes of the DLC meeting held on 13.01.2021 Order No. 1124/2020/KSD/KDP (572) Dated: 30.01.2021 As per the Government order read as 1st paper above, Government have constituted an Empowered Committee for the implementation of Dr. P. Prabhakaran Commission Report on Kasaragod Development Package. An amount of Rs.75.00 crore under the Head of Account 2551-60-101-97 has been earmarked during the financial year 2020-21 for implementing the projects included in the Kasaragod Development Package. As per the Government order read as 2nd paper above, a District Level Committee was constituted for granting administrative sanction for the projects below Rs. 5.00 crore under Kasaragod Development Package, based on the recommendation of State Level Empowered Committee. As per the letter read as 3rd paper above, the Executive Engineer, Minor Irrigation Division, Kasaragod furnished the D.P.R of the project “Integrated water management of Kasaragod district- Renovation of VCB at Edaneer across Mogral river in Chengala G.P” for an amount of Rs.