(12) Patent Application Publication (10) Pub. No.: US 2015/0259700 A1 Elling Et Al
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

From the Ground up the First Fifty Years of Mccain Foods
CHAPTER TITLE i From the Ground up the FirSt FiFty yearS oF mcCain FoodS daniel StoFFman In collaboratI on wI th t ony van l eersum ii FROM THE GROUND UP CHAPTER TITLE iii ContentS Produced on the occasion of its 50th anniversary Copyright © McCain Foods Limited 2007 Foreword by Wallace McCain / x by All rights reserved. No part of this book, including images, illustrations, photographs, mcCain FoodS limited logos, text, etc. may be reproduced, modified, copied or transmitted in any form or used BCE Place for commercial purposes without the prior written permission of McCain Foods Limited, Preface by Janice Wismer / xii 181 Bay Street, Suite 3600 or, in the case of reprographic copying, a license from Access Copyright, the Canadian Toronto, Ontario, Canada Copyright Licensing Agency, One Yonge Street, Suite 1900, Toronto, Ontario, M6B 3A9. M5J 2T3 Chapter One the beGinninG / 1 www.mccain.com 416-955-1700 LIBRARY AND ARCHIVES CANADA CATALOGUING IN PUBLICATION Stoffman, Daniel Chapter Two CroSSinG the atlantiC / 39 From the ground up : the first fifty years of McCain Foods / Daniel Stoffman For copies of this book, please contact: in collaboration with Tony van Leersum. McCain Foods Limited, Chapter Three aCroSS the Channel / 69 Director, Communications, Includes index. at [email protected] ISBN: 978-0-9783720-0-2 Chapter Four down under / 103 or at the address above 1. McCain Foods Limited – History. 2. McCain, Wallace, 1930– . 3. McCain, H. Harrison, 1927–2004. I. Van Leersum, Tony, 1935– . II. McCain Foods Limited Chapter Five the home Front / 125 This book was printed on paper containing III. -

US20200383331A1.Pdf
US 20200383331A1 IN (19United States ( 12 ) Patent Application Publication ( Pub. No.:USQO2Q/QZ8333l Al HEINRICHER ( 43 ) Pub . Date : Dec. 10 , 2020 ( 54 ) COMPOSITIONS AND METHODS FOR AOIN 43/40 ( WQOQI LARGE - SCALE IN VITRO PLANT AOIN 43/08 ( 2006.01 ) BIOCULTURE A01N 37/52 ( 2006.01 ) AOIN 4730 ( 2006.01 ) ( II ) Applicant: BQOSHIQOQT LLC , Hailey, IDUS A016 22/15 ( WQGOI ( 52 ) U.S. CI . ( 72 ) Inventor: Jackie HEINRICHER , Anacortes , WA CPC AOIN 43/90 ( 2013.01 ) ; AO1G 31/00 (US ) ( 2013.01 ) ; A01N 59/08 ( 2013.01 ) ; A01N 59/20 ( 2013.01 ) ; A01N 59/16 ( 2013.01 ) ; ( 21 ) Appl . No .: 16 /728,478 A01N 59/14 ( 2013.01 ) ; A01N 31/06 ( 2013.01 ) ; A01N 43/78 ( 2013.01 ) ; A01N ( 22 ) Filed : Dec. 27 , 2019 37/10 ( 2013.01 ) ; A01N 43/82 ( 2013.01 ) ; AOIN 59/12 ( 2013.01 ) ; AOIN 37/44 Related U.S. Application Data ( 2013.01 ) ; A01N 43/40 ( 2013.01 ) ; A01N ( 63 ) Continuation of application No. PCT /US2018 / 43/08 ( 2013.01 ) ; A01N 37/52 ( 2013.01 ) ; 040637 , filed on Jul. 2 , 2018 , Continuation of appli AOIN 47/30 ( 2013.01 ) ; A01G 22/15 cation No. PCT/ US2018 / 040646 , filed on Jul. 2 , ( 2018.02 ) ; A01N 59/00 ( 2013.01 ) 2018 . ( 60 ) Provisional application No. 62 / 527,946 , filed on Jun . ( 57 ) ABSTRACT 3Q , provisional application No. 62 /6II , & a , The present invention provides media , kits , systems , and filed on Dec. 29 , 2017 , provisional application No. methods for achieving large scale pistachio production 62 / 527,862 , filed on Jun . 30 , 2017 . within a short time via bioculture , large scale yam produc tion within a short time via bioculture, high multiplication Publication Classification rate of plants including cannabis via in vitro micropropaga ( 51 ) Int . -

International Union for the Protection of New Varieties of Plants Geneva
E TG/23/6 ORIGINAL: English DATE: 2004-03-31 INTERNATIONAL UNION FOR THE PROTECTION OF NEW VARIETIES OF PLANTS GENEVA * POTATO (Solanum tuberosum L.) GUIDELINES FOR THE CONDUCT OF TESTS FOR DISTINCTNESS, UNIFORMITY AND STABILITY Alternative Names: * Latin English French German Spanish Solanum tuberosum L., Potato Pomme de terre Kartoffel Papa, Patata S. tuberosum L. sensu lato ASSOCIATED DOCUMENTS These guidelines should be read in conjunction with document TG/1/3, “G eneral Introduction to the Examination of Distinctness, Uniformity and Stability and the Development of Harmonized Descriptions of New Varieties of Plants” (hereinafter referred to as the “General Introduction”) and its associated “TGP” documents. * These names were correct at the time of the introduction of these Test Guidelines but may be revised or updated. [Readers are advised to consult the UPOV Code, which can be found on the UPOV Website (www.upov.int), for the latest infor mation.] TG/23/6 Potato, 2004 -03 -31 - 2 - TABLE OF CONTENTS 1. SUBJECT OF THESE TES T GUIDELINES ................................ ................................ ................................ .. 3 2. MATERIAL REQUIRED ................................ ................................ ................................ ............................... 3 3. METHOD OF EXAMINATIO N................................ ................................ ................................ ..................... 3 3.1 Duration of Tests ................................ ................................ ............................... -

Report of a Working Group on Potato: First Meeting, 23-25 March 2000
European Cooperative Programme for Crop Genetic Report Resources Networks ECP GR of a Working Group on Potato First Meeting 23–25 March 2000, Wageningen, The Netherlands R. Hoekstra, L. Maggioni and E. Lipman, compilers <www.futureharvest.org> IPGRI is a Future Harvest Centre supported by the Consultative Group on International Agricultural Research (CGIAR) Report ECP GR of a working group on Potato First Meeting 23–25 March 2000, Wageningen, The Netherlands R. Hoekstra, L. Maggioni and E. Lipman, compilers The International Plant Genetic Resources Institute (IPGRI) is an autonomous international scientific organization, supported by the Consultative Group on International Agricultural Research (CGIAR). IPGRI's mandate is to advance the conservation and use of genetic diversity for the well-being of present and future generations. IPGRI's headquarters is based in Maccarese, near Rome, Italy, with offices in another 19 countries worldwide. The Institute operates through three programmes: (1) the Plant Genetic Resources Programme, (2) the CGIAR Genetic Resources Support Programme and (3) the International Network for the Improvement of Banana and Plantain (INIBAP). The international status of IPGRI is conferred under an Establishment Agreement which, by January 2001, had been signed and ratified by the Governments of Algeria, Australia, Belgium, Benin, Bolivia, Brazil, Burkina Faso, Cameroon, Chile, China, Congo, Costa Rica, Côte d’Ivoire, Cyprus, Czech Republic, Denmark, Ecuador, Egypt, Greece, Guinea, Hungary, India, Indonesia, Iran, Israel, Italy, Jordan, Kenya, Malaysia, Mauritania, Morocco, Norway, Pakistan, Panama, Peru, Poland, Portugal, Romania, Russia, Senegal, Slovakia, Sudan, Switzerland, Syria, Tunisia, Turkey, Uganda and Ukraine. In 2000 financial support for the Research Agenda of IPGRI was provided by the Governments of Armenia, Australia, Austria, Belgium, Brazil, Bulgaria, Canada, China, Croatia, Cyprus, Czech Republic, Denmark, Estonia, F.R. -
2012-USA Seed by Variety Accepted
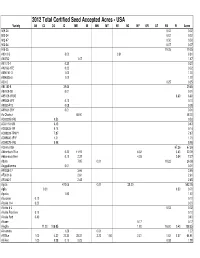
2012 Total Certified Seed Accepted Acres - USA Variety AK CA CO ID ME MI MN MT NE ND NY OR UT WA WI Acres 509-24 0.02 0.02 802-34 0.02 0.02 902-47 0.50 0.50 903-34 0.07 0.07 915-25 19.25 19.25 A0012-5 3.00 0.31 3.31 A00732 1.47 1.47 A01010-1 0.20 0.20 A02062-1TE 0.22 0.22 A096141-3 1.00 1.00 A096305-3 1.00 1.00 A2012 0.25 0.25 A84180-8 39.65 39.65 A87009-28 0.01 0.01 A95109-1RUS 6.60 6.60 A99326-4PY 0.10 0.10 A9932-PY2 0.08 0.08 A99331-2RY 0.01 0.01 Ac Chaleur 48.00 48.00 AC00395-2RU 0.53 0.53 AC01151-5W 0.43 0.43 AC03433-1W 0.14 0.14 AC99329-7PW/Y 7.87 7.87 AC99330-1P/Y 1.71 1.71 AC99375-1RU 5.98 5.98 Accumulator 67.26 67.26 Adirondack Blue 0.20 11.97 6.32 3.60 22.09 Adirondack Red 0.10 2.37 4.26 0.54 7.27 Adora 7.85 0.01 18.22 26.08 Aeggeblomme 0.01 0.01 AF0338-17 3.66 3.66 AF3001-6 2.61 2.61 AF3362-1 2.45 2.45 Agata 473.05 0.01 29.00 502.06 Agila 0.20 0.50 0.70 Agusta 1.50 1.50 Alasclear 0.10 0.10 Alaska 114 0.20 0.20 Alaska 8-3 0.02 0.02 Alaska Frostless 0.10 0.10 Alaska Red 0.40 0.40 Albane 0.17 0.17 Alegria 11.20 108.85 1.90 16.00 0.40 138.35 Alexandra 1.26 0.01 1.27 All Blue 1.00 4.32 22.30 28.37 3.22 1.50 2.31 1.52 0.37 64.91 All Red 1.00 0.28 0.10 0.23 0.38 1.99 Variety AK CA CO ID ME MI MN MT NE ND NY OR UT WA WI Acres Allagash 1.00 1.00 Allian 91.74 0.01 91.75 Almera 0.37 0.37 Alpine Russet 158.00 101.01 60.10 6.21 73.58 19.00 13.85 431.75 Alta Crown 7.00 7.00 Alturas 1935.05 0.01742.15 161.00 2838.21 Amandine 0.24 0.24 Amarosa 2.00 0.20 0.01 0.03 0.64 7.60 10.48 Ambra 0.40 3.84 4.51 8.75 Amey 1.23 1.23 Ampera 17.64 0.01 17.65 -

Potato - Wikipedia, the Free Encyclopedia
Potato - Wikipedia, the free encyclopedia Log in / create account Article Talk Read View source View history Our updated Terms of Use will become effective on May 25, 2012. Find out more. Main page Potato Contents From Wikipedia, the free encyclopedia Featured content Current events "Irish potato" redirects here. For the confectionery, see Irish potato candy. Random article For other uses, see Potato (disambiguation). Donate to Wikipedia The potato is a starchy, tuberous crop from the perennial Solanum tuberosum Interaction of the Solanaceae family (also known as the nightshades). The word potato may Potato Help refer to the plant itself as well as the edible tuber. In the region of the Andes, About Wikipedia there are some other closely related cultivated potato species. Potatoes were Community portal first introduced outside the Andes region four centuries ago, and have become Recent changes an integral part of much of the world's cuisine. It is the world's fourth-largest Contact Wikipedia food crop, following rice, wheat and maize.[1] Long-term storage of potatoes Toolbox requires specialised care in cold warehouses.[2] Print/export Wild potato species occur throughout the Americas, from the United States to [3] Uruguay. The potato was originally believed to have been domesticated Potato cultivars appear in a huge variety of [4] Languages independently in multiple locations, but later genetic testing of the wide variety colors, shapes, and sizes Afrikaans of cultivars and wild species proved a single origin for potatoes in the area -

Tesis Doctoral 2017
MEJORA GENÉTICA DE PATATA PARA COMPUESTOS BIOACTIVOS Y CAPACIDAD ANTIOXIDANTE Roberto Tierno Fernández Tesis doctoral 2017 MEJORA GENÉTICA DE PATATA PARA COMPUESTOS BIOACTIVOS Y CAPACIDAD ANTIOXIDANTE Roberto Tierno Fernández Tesis doctoral Director: Dr. D. José Ignacio Ruiz de Galarreta Tutora: Dra. Dª. Mª Teresa Lacuesta Calvo (c)2017 ROBERTO TIERNO FERNANDEZ AGRADECIMIENTOS Este trabajo no hubiera sido posible sin la participación de muchos compañeros que, en mayor o menor medida, han contribuido a la realización de esta tesis. En primer lugar, mi reconocimiento al director de tesis, José Ignacio Ruiz de Galarreta, que apostó por mí desde el primer momento y durante este tiempo, me ha dado toda la confianza, libertad y apoyo que he necesitado. A la dirección de Neiker, por haberme acogido durante estos cuatro años y al INIA, organismo que financió este proyecto y posibilitó mi formación. Al Departamento de Biología Vegetal y Ecología de la UPV-EHU, por permitirme presentar este trabajo y especialmente a la Dra. Mª Teresa Lacuesta Calvo por aceptar ser tutora de esta tesis. Quiero agradecer la paciencia y buena disposición de muchas personas sin cuyas enseñanzas y consejos este trabajo jamás hubiera podido realizarse. Entre ellas, quiero hacer una mención muy especial a Isi, Carlos Castaño, Carlos Herrán y Bego. Otras personas con las que he tenido la oportunidad de trabajar y formarme son Ainara, Carmen, Silvia, Patrick Riga, Leire, Berdaitz y Jon. Por supuesto, quiero y debo reconocer también el inestimable aporte de Néstor, Raquel López, Mikel González, Emma López de Armentia, Jon Lemos y otros camaradas becarios, cuya guía, consejo y conocimiento ha sido determinante. -
Seed Potato Directory 2017
The farm operation grows 93 acres of field generations one and two seed, operates 4 greenhouses producing conventional and NFT minitubers. Our stewardship of this seed continues through WISCONSIN the certification Our of stewardship these seed oflots this on seed Wisconsin continues seed through grower t farms, there is no other program like it. CERTIFIED The program maintains variety trueness to type; selecting and testing clones, rogueing of weak, genetic variants, and diseased plants to continue to develop and maintain germplasm of your SEED POTATOES favorite varieties at our laboratory. 103 Years of Seed Growing Tradition A Century Long Tradition Pioneers In Seed Potato Certification Administered since inception by the College of Agricultural and Life Sciences, University of Wisconsin – Madison, the program Much of the early research work on potato diseases and how retains a full-time staff of experienced professionals to ensure they spread was done Scientists in Germany found and that, Holland through around careful the monitoring turn thoroughness and impartiality in inspection and certification of the century. Scientists found that, through careful monitoring procedures. o of the crop and removal of unhealthy plants, Similar they could research maintain soon was a vigorous, healthy stock indefinitely. Similar research soon was Through providing information, exercising technical skill, doing b being conducted in the United States. research directed at solving problems, and conducting outreach activities, the University meets the growers at the field level. USDA plant pathologist W.A. Orton had studied potato This special relationship to the academic community brings new certification in Germany and upon his return, began to work with T information on pathogens, best practices, and introduces high potato growers and Universities to introduce those concepts quality basic seed into the marketplace. -

Starch and Antioxidant Properties of Quebec-Bred Potato Genotypes
Starch and Antioxidant Properties of Quebec-bred Potato Genotypes Christina Larder Department of Plant Science Macdonald Campus of McGill University Montréal, Québec, Canada December 2015 A thesis submitted to McGill University in partial fulfillment of the requirements for the degree of Master of Science ©Christina Larder, 2015 I Table of Contents Abstract ........................................................................................................................................ VII Résumé ....................................................................................................................................... VIII Acknowledgements ........................................................................................................................ X Contribution of Authors ................................................................................................................ XI List of Tables ............................................................................................................................... XII List of Figures .............................................................................................................................. XV List of Abbreviations ................................................................................................................. XVI Chapter 1: General Introduction ..................................................................................................... 1 1.1 Introduction .......................................................................................................................... -

Foodservice Product Guide Draft
Page 1 Seasonal Calendar Seasons subject to weather Code Product Description Usual Case Size Other Units of Sale Special Notes Storage Temp Jan Feb Mar Apr May Jun Jul Aug Sep Oct Nov Dec = usually available UK = UK season available = not available Please note seasons can vary year to year FRUIT APPBRAE Apple Braeburn Box (12.5kg) kg, each C UK UK UK UK UK APPBRAM Apple Bramley Box (12.5kg) kg, each C UK UK UK UK UK UK UK UK UK UK UK UK APPCOX Apple Coxes Box (12.5kg) kg, each C UK UK UK UK UK UK UK UK APPDIS Apple Discovery Box (12.5kg) kg, each C UK UK APPDELE Apple Delbard Estive Box (12.5kg) kg, each C UK UK UK APPLEJFUJI Apple Fuji Box (12.5kg) kg, each 24hr notice C APPGD Apple Golden Delicious Box (12.5kg) kg, each C APPGS Apple Granny Smith Box (18kg / 12.5kg) kg, each C APPJAZ Apple Jazz Box (12.5kg) kg, each C UK UK UK UK UK UK APPPL Apple Pink Lady Box (12.5kg) kg, each C APPRD Apple Red Delicious Box (18kg / 12.5kg) kg, each C APPREDW Apple Red Washington Box (12.5kg) kg, each C APPRG Applw Royal Gala Box (12.5kg) kg, each C UK UK UK UK UK UK UK APPRUS Apple Russet Box (12.5kg) kg, each C UK UK UK UK UK UK UK APPCRAB Apple Crab 500g punnet 24hr notice C UK UK APPCUS Apples Custard Box (12kg) box only 24hr notice C APRI Apricots Box (5kg) kg, each C AVOH Avocado Haas Box (Count 16) each C BABYKIW Baby Kiwi 125g punnet 24hours C BANB Baby Banana Box (3kg) kg, bunches C PLANTGR Banana Green Plantain Box (15kg) kg min order 5kg C BANL Banana Leaves 2 x 500g 500g packet C BAN Banana Med/Large Box (18kg) kg, each A BUDHA Budhas -

Potatoes in Practice 2012 Event Guide
3RWDWRHV3RWDWRHVPotatoes LQ3UDFWLFHLQ3UDFWLFHin Practice 2012 ThursdayThursday 12th 12th9th August August 9.30am9.30am to to 4.30pm 4.30pm4.30pm BalrudderyBalruddery Farm Farm InvergowrieInvergowrieInvergowrie DundeeDundee DD2 DD2DD2 5LJ 5LJ5LJ SupportedSupported by by FieldField Trials Trials & && Demonstrations, Demonstrations,Demonstrations, SeminarsSeminars and and Exhibitors Exhibitors Guide Guide ProgrammeContents Welcome to Potatoes in Practice 2012 Welcome to Potatoes in Practice 2012.................................................... 1 The James Hutton Institute, SAC, Agrii (formerly Masstock Arable) and the Potato Programme ................................................................................................ 2 Council welcome you to Potatoes in Practice 2012. This event is Britain’s premier field based event dedicated to the potato industry, attracting more international Site Plan ..................................................................................................... 4 visitors each year. Seminars .................................................................................................... 6 This is the fourth year PiP has been held at the James Hutton Institute’s Balruddery Farm. The event is supported by Potato Review. Exhibitor List .............................................................................................. 8 This is a unique opportunity for farmers, advisers and others to view government and industry- Field Trials and Demonstrations ........................................................... -

E:\Jobs\36000-36999\36289P SSPGA\Originals\36289 Cover.Wpd
2006 - 2007 Saskatchewan Seed Potato Directory SSPGA BOARD OF DIRECTORS & STAFF Elected Directors Brad Bartko, President Scott Anderson, Vice-President Bill Ingram, Treasurer Elly Könst Jacob Vanderschaaf Ex-Officio Directors Dr. Doug Waterer, University of Saskatchewan Connie Achtymichuk, Saskatchewan Agriculture, Food & Rural Revitalization Dr. Jazeem Wahab, Canada-Saskatchewan Irrigation Diversification Centre Staff Linda Sinclair Address Box 386 Outlook, Saskatchewan Canada S0L 2N0 Tel.: 306-867-2078 Fax: 306-867-2102 E-mail: [email protected] Website: http://sasktelwebsite.net/sspga 2006 - 2007 SASKATCHEWAN SEED POTATO DIRECTORY This Seed Directory is published by : Saskatchewan Seed Potato Growers’ Association Inc. © 2006 Printed in Canada TABLE OF CONTENTS SSPGA Mandate .............................. 1 Equivalency Tables ............................ 3 NORTHERN VIGOR® .......................... 5 Benefits of Buying Saskatchewan Seed ............. 7 Canadian Certification Program ................... 8 Seed Identification ............................ 10 Procedures to Control Bacterial Ring Rot ........... 11 Post-Harvest Testing .......................... 12 Tuber Quality Standards ........................ 13 Notice to Buyer ............................... 14 Acreage Listing by Variety ...................... 15 Section 1 - Nuclear to Elite 2 ................. 16 Section 2 - Elite 3 to Certified ................. 21 Section 3 - Private Varieties .................. 23 List of Members, 2006 - 2007 ................... 24 SSPGA MANDATE