Ser 15 of WEE1B Is a Potential PKA Phosphorylation Target in G2/M Transition in One-Cell Stage Mouse Embryos
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplemental Information to Mammadova-Bach Et Al., “Laminin Α1 Orchestrates VEGFA Functions in the Ecosystem of Colorectal Carcinogenesis”
Supplemental information to Mammadova-Bach et al., “Laminin α1 orchestrates VEGFA functions in the ecosystem of colorectal carcinogenesis” Supplemental material and methods Cloning of the villin-LMα1 vector The plasmid pBS-villin-promoter containing the 3.5 Kb of the murine villin promoter, the first non coding exon, 5.5 kb of the first intron and 15 nucleotides of the second villin exon, was generated by S. Robine (Institut Curie, Paris, France). The EcoRI site in the multi cloning site was destroyed by fill in ligation with T4 polymerase according to the manufacturer`s instructions (New England Biolabs, Ozyme, Saint Quentin en Yvelines, France). Site directed mutagenesis (GeneEditor in vitro Site-Directed Mutagenesis system, Promega, Charbonnières-les-Bains, France) was then used to introduce a BsiWI site before the start codon of the villin coding sequence using the 5’ phosphorylated primer: 5’CCTTCTCCTCTAGGCTCGCGTACGATGACGTCGGACTTGCGG3’. A double strand annealed oligonucleotide, 5’GGCCGGACGCGTGAATTCGTCGACGC3’ and 5’GGCCGCGTCGACGAATTCACGC GTCC3’ containing restriction site for MluI, EcoRI and SalI were inserted in the NotI site (present in the multi cloning site), generating the plasmid pBS-villin-promoter-MES. The SV40 polyA region of the pEGFP plasmid (Clontech, Ozyme, Saint Quentin Yvelines, France) was amplified by PCR using primers 5’GGCGCCTCTAGATCATAATCAGCCATA3’ and 5’GGCGCCCTTAAGATACATTGATGAGTT3’ before subcloning into the pGEMTeasy vector (Promega, Charbonnières-les-Bains, France). After EcoRI digestion, the SV40 polyA fragment was purified with the NucleoSpin Extract II kit (Machery-Nagel, Hoerdt, France) and then subcloned into the EcoRI site of the plasmid pBS-villin-promoter-MES. Site directed mutagenesis was used to introduce a BsiWI site (5’ phosphorylated AGCGCAGGGAGCGGCGGCCGTACGATGCGCGGCAGCGGCACG3’) before the initiation codon and a MluI site (5’ phosphorylated 1 CCCGGGCCTGAGCCCTAAACGCGTGCCAGCCTCTGCCCTTGG3’) after the stop codon in the full length cDNA coding for the mouse LMα1 in the pCIS vector (kindly provided by P. -

Gene Symbol Gene Description ACVR1B Activin a Receptor, Type IB
Table S1. Kinase clones included in human kinase cDNA library for yeast two-hybrid screening Gene Symbol Gene Description ACVR1B activin A receptor, type IB ADCK2 aarF domain containing kinase 2 ADCK4 aarF domain containing kinase 4 AGK multiple substrate lipid kinase;MULK AK1 adenylate kinase 1 AK3 adenylate kinase 3 like 1 AK3L1 adenylate kinase 3 ALDH18A1 aldehyde dehydrogenase 18 family, member A1;ALDH18A1 ALK anaplastic lymphoma kinase (Ki-1) ALPK1 alpha-kinase 1 ALPK2 alpha-kinase 2 AMHR2 anti-Mullerian hormone receptor, type II ARAF v-raf murine sarcoma 3611 viral oncogene homolog 1 ARSG arylsulfatase G;ARSG AURKB aurora kinase B AURKC aurora kinase C BCKDK branched chain alpha-ketoacid dehydrogenase kinase BMPR1A bone morphogenetic protein receptor, type IA BMPR2 bone morphogenetic protein receptor, type II (serine/threonine kinase) BRAF v-raf murine sarcoma viral oncogene homolog B1 BRD3 bromodomain containing 3 BRD4 bromodomain containing 4 BTK Bruton agammaglobulinemia tyrosine kinase BUB1 BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) BUB1B BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) C9orf98 chromosome 9 open reading frame 98;C9orf98 CABC1 chaperone, ABC1 activity of bc1 complex like (S. pombe) CALM1 calmodulin 1 (phosphorylase kinase, delta) CALM2 calmodulin 2 (phosphorylase kinase, delta) CALM3 calmodulin 3 (phosphorylase kinase, delta) CAMK1 calcium/calmodulin-dependent protein kinase I CAMK2A calcium/calmodulin-dependent protein kinase (CaM kinase) II alpha CAMK2B calcium/calmodulin-dependent -

Tumour-Agnostic Therapy for Pancreatic Cancer and Biliary Tract Cancer
diagnostics Review Tumour-Agnostic Therapy for Pancreatic Cancer and Biliary Tract Cancer Shunsuke Kato Department of Clinical Oncology, Juntendo University Graduate School of Medicine, 2-1-1, Hongo, Bunkyo-ku, Tokyo 113-8421, Japan; [email protected]; Tel.: +81-3-5802-1543 Abstract: The prognosis of patients with solid tumours has remarkably improved with the develop- ment of molecular-targeted drugs and immune checkpoint inhibitors. However, the improvements in the prognosis of pancreatic cancer and biliary tract cancer is delayed compared to other carcinomas, and the 5-year survival rates of distal-stage disease are approximately 10 and 20%, respectively. How- ever, a comprehensive analysis of tumour cells using The Cancer Genome Atlas (TCGA) project has led to the identification of various driver mutations. Evidently, few mutations exist across organs, and basket trials targeting driver mutations regardless of the primary organ are being actively conducted. Such basket trials not only focus on the gate keeper-type oncogene mutations, such as HER2 and BRAF, but also focus on the caretaker-type tumour suppressor genes, such as BRCA1/2, mismatch repair-related genes, which cause hereditary cancer syndrome. As oncogene panel testing is a vital approach in routine practice, clinicians should devise a strategy for improved understanding of the cancer genome. Here, the gene mutation profiles of pancreatic cancer and biliary tract cancer have been outlined and the current status of tumour-agnostic therapy in these cancers has been reported. Keywords: pancreatic cancer; biliary tract cancer; targeted therapy; solid tumours; driver mutations; agonist therapy Citation: Kato, S. Tumour-Agnostic Therapy for Pancreatic Cancer and 1. -

Systematic Screening for Potential Therapeutic Targets in Osteosarcoma Through a Kinome-Wide CRISPR-Cas9 Library
Cancer Biol Med 2020. doi: 10.20892/j.issn.2095-3941.2020.0162 ORIGINAL ARTICLE Systematic screening for potential therapeutic targets in osteosarcoma through a kinome-wide CRISPR-Cas9 library Yuanzhong Wu*, Liwen Zhou*, Zifeng Wang, Xin Wang, Ruhua Zhang, Lisi Zheng, Tiebang Kang Sun Yat-sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangzhou 510060, China ABSTRACT Objective: Osteosarcoma is the most common primary malignant bone tumor. However, the survival of patients with osteosarcoma has remained unchanged during the past 30 years, owing to a lack of efficient therapeutic targets. Methods: We constructed a kinome-targeting CRISPR-Cas9 library containing 507 kinases and 100 nontargeting controls and screened the potential kinase targets in osteosarcoma. The CRISPR screening sequencing data were analyzed with the Model-based Analysis of Genome-wide CRISPR/Cas9 Knockout (MAGeCK) Python package. The functional data were applied in the 143B cell line through lenti-CRISPR-mediated gene knockout. The clinical significance of kinases in the survival of patients with osteosarcoma was analyzed in the R2: Genomics Analysis and Visualization Platform. Results: We identified 53 potential kinase targets in osteosarcoma. Among these targets, we analyzed 3 kinases, TRRAP, PKMYT1, and TP53RK, to validate their oncogenic functions in osteosarcoma. PKMYT1 and TP53RK showed higher expression in osteosarcoma than in normal bone tissue, whereas TRRAP showed no significant difference. High expression of all 3 kinases was associated with relatively poor prognosis in patients with osteosarcoma. Conclusions: Our results not only offer potential therapeutic kinase targets in osteosarcoma but also provide a paradigm for functional genetic screening by using a CRISPR-Cas9 library, including target design, library construction, screening workflow, data analysis, and functional validation. -

DNAJB1–PRKACA Fusion Kinase Interacts with Β-Catenin and the Liver
DNAJB1–PRKACA fusion kinase interacts with INAUGURAL ARTICLE β-catenin and the liver regenerative response to drive fibrolamellar hepatocellular carcinoma Edward R. Kastenhubera,b, Gadi Lalazarc, Shauna L. Houlihana, Darjus F. Tschaharganehd,e, Timour Baslana, Chi-Chao Chena, David Requenac, Sha Tiana, Benedikt Bosbachf, John E. Wilkinsong, Sanford M. Simonc, and Scott W. Lowea,h,1 aDepartment of Cancer Biology and Genetics, Memorial Sloan Kettering Cancer Center, New York, NY 10065; bLouis V. Gerstner Jr. Graduate School of Biomedical Sciences, Memorial Sloan Kettering Cancer Center, New York, NY 10065; cLaboratory of Cellular Biophysics, The Rockefeller University, New York, NY 10065; dHelmholtz University Group “Cell Plasticity and Epigenetic Remodeling,” German Cancer Research Center (DKFZ), 69120 Heidelberg, Germany; eInstitute of Pathology, University Hospital, 69120 Heidelberg, Germany; fOncology Target Discovery Program, Pfizer Inc., Pearl River, NY 10965; gDepartment of Pathology, University of Michigan School of Medicine, Ann Arbor, MI 48109; and hHoward Hughes Medical Institute, New York, NY 10065 This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected in 2017. Contributed by Scott W. Lowe, October 26, 2017 (sent for review September 22, 2017; reviewed by Nabeel M. Bardeesy and David A. Largaespada) A segmental deletion resulting in DNAJB1–PRKACA gene fusion is any known etiological risk factors such as alcoholism, chronic hep- now recognized as the signature genetic event of fibrolamellar hepa- atitis infection, or liver flukes (8). tocellular carcinoma (FL-HCC), a rare but lethal liver cancer that pri- Currently, FL-HCC is diagnosed on the basis of histological marily affects adolescents and young adults. -

The Combination of the PARP Inhibitor Olaparib and the Wee1 Inhibitor AZD1775 As a New Therapeutic Option for Small Cell Lung Cancer
Author Manuscript Published OnlineFirst on June 25, 2018; DOI: 10.1158/1078-0432.CCR-17-2805 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. The combination of the PARP inhibitor olaparib and the Wee1 inhibitor AZD1775 as a new therapeutic option for small cell lung cancer Alice Lallo1,*, Kristopher K Frese1,*, Christopher J Morrow1, Robert Sloane1, Sakshi Gulati1, Maximillian W Schenk1, Francesca Trapani1, Nicole Simms1, Melanie Galvin1, Stewart Brown1, Cassandra L Hodgkinson1, Lynsey Priest1, Adina Hughes4, Zhongwu Lai5, Elaine Cadogan4, Garima Khandelwal3, Kathryn L Simpson1, Crispin Miller3, Fiona Blackhall2, Mark J O’Connor4,#, and Caroline Dive1,# 1 Clinical and Experimental Pharmacology Group, Cancer Research UK Manchester Institute, University of Manchester, Manchester, M20 4BX, UK. 2 Institute of Cancer Sciences, University of Manchester, Manchester, United Kingdom. Christie NHS Foundation Trust, Manchester, M20 4BX, UK. 3 RNA Biology Group, Cancer Research UK Manchester Institute, University of Manchester, Manchester, M20 4BX, UK. 4 Oncology Innovative Medicines and Early Development Biotech Unit, AstraZeneca, Cambridge, CB2 0RE, UK. 5 Oncology Innovative Medicines and Early Development Biotech Unit, AstraZeneca, Waltham, 02451, USA. *These authors contributed equally to this work Running title: PARP and Wee1 inhibition in patient-derived models of SCLC C. Dive receives funding from the Cancer Research UK Manchester Institute core (C5759/A27412), the Cancer Research UK Manchester -

Bioinformatics-Based Screening of Key Genes for Transformation of Liver
Jiang et al. J Transl Med (2020) 18:40 https://doi.org/10.1186/s12967-020-02229-8 Journal of Translational Medicine RESEARCH Open Access Bioinformatics-based screening of key genes for transformation of liver cirrhosis to hepatocellular carcinoma Chen Hao Jiang1,2, Xin Yuan1,2, Jiang Fen Li1,2, Yu Fang Xie1,2, An Zhi Zhang1,2, Xue Li Wang1,2, Lan Yang1,2, Chun Xia Liu1,2, Wei Hua Liang1,2, Li Juan Pang1,2, Hong Zou1,2, Xiao Bin Cui1,2, Xi Hua Shen1,2, Yan Qi1,2, Jin Fang Jiang1,2, Wen Yi Gu4, Feng Li1,2,3 and Jian Ming Hu1,2* Abstract Background: Hepatocellular carcinoma (HCC) is the most common type of liver tumour, and is closely related to liver cirrhosis. Previous studies have focussed on the pathogenesis of liver cirrhosis developing into HCC, but the molecular mechanism remains unclear. The aims of the present study were to identify key genes related to the transformation of cirrhosis into HCC, and explore the associated molecular mechanisms. Methods: GSE89377, GSE17548, GSE63898 and GSE54236 mRNA microarray datasets from Gene Expression Omni- bus (GEO) were analysed to obtain diferentially expressed genes (DEGs) between HCC and liver cirrhosis tissues, and network analysis of protein–protein interactions (PPIs) was carried out. String and Cytoscape were used to analyse modules and identify hub genes, Kaplan–Meier Plotter and Oncomine databases were used to explore relationships between hub genes and disease occurrence, development and prognosis of HCC, and the molecular mechanism of the main hub gene was probed using Kyoto Encyclopedia of Genes and Genomes(KEGG) pathway analysis. -

PRKACA Mediates Resistance to HER2-Targeted Therapy in Breast Cancer Cells and Restores Anti-Apoptotic Signaling
Oncogene (2015) 34, 2061–2071 © 2015 Macmillan Publishers Limited All rights reserved 0950-9232/15 www.nature.com/onc ORIGINAL ARTICLE PRKACA mediates resistance to HER2-targeted therapy in breast cancer cells and restores anti-apoptotic signaling SE Moody1,2,3, AC Schinzel1, S Singh1, F Izzo1, MR Strickland1, L Luo1,2, SR Thomas3, JS Boehm3, SY Kim4, ZC Wang5,6 and WC Hahn1,2,3 Targeting HER2 with antibodies or small molecule inhibitors in HER2-positive breast cancer leads to improved survival, but resistance is a common clinical problem. To uncover novel mechanisms of resistance to anti-HER2 therapy in breast cancer, we performed a kinase open reading frame screen to identify genes that rescue HER2-amplified breast cancer cells from HER2 inhibition or suppression. In addition to multiple members of the MAPK (mitogen-activated protein kinase) and PI3K (phosphoinositide 3-kinase) signaling pathways, we discovered that expression of the survival kinases PRKACA and PIM1 rescued cells from anti-HER2 therapy. Furthermore, we observed elevated PRKACA expression in trastuzumab-resistant breast cancer samples, indicating that this pathway is activated in breast cancers that are clinically resistant to trastuzumab-containing therapy. We found that neither PRKACA nor PIM1 restored MAPK or PI3K activation after lapatinib or trastuzumab treatment, but rather inactivated the pro-apoptotic protein BAD, the BCl-2-associated death promoter, thereby permitting survival signaling through BCL- XL. Pharmacological blockade of BCL-XL/BCL-2 partially abrogated the rescue effects conferred by PRKACA and PIM1, and sensitized cells to lapatinib treatment. These observations suggest that combined targeting of HER2 and the BCL-XL/BCL-2 anti-apoptotic pathway may increase responses to anti-HER2 therapy in breast cancer and decrease the emergence of resistant disease. -

Personalized Prediction of Acquired Resistance to EGFR-Targeted Inhibitors Using a Pathway-Based Machine Learning Approach
Cancers 2019, 11, x S1 of S9 Supplementary Materials: Personalized Prediction of Acquired Resistance to EGFR-Targeted Inhibitors Using a Pathway-Based Machine Learning Approach Young Rae Kim, Yong Wan Kim, Suh Eun Lee, Hye Won Yang and Sung Young Kim Table S1. Characteristics of individual studies. Sample Size Origin of Cancer Drug Dataset Platform S AR (Cell Lines) Lung cancer Agilent-014850 Whole Human GSE34228 26 26 (PC9) Genome Microarray 4x44K Gefitinib Epidermoid carcinoma Affymetrix Human Genome U133 GSE10696 3 3 (A431) Plus 2.0 Head and neck cancer Illumina HumanHT-12 V4.0 GSE62061 12 12 (Cal-27, SSC-25, FaDu, expression beadchip SQ20B) Erlotinib Head and neck cancer Illumina HumanHT-12 V4.0 GSE49135 3 3 (HN5) expression beadchip Lung cancer (HCC827, Illumina HumanHT-12 V3.0 GSE38310 3 6 ER3, T15-2) expression beadchip Lung cancer Illumina HumanHT-12 V3.0 GSE62504 1 2 (HCC827) expression beadchip Afatinib Lung cancer * Illumina HumanHT-12 V4.0 GSE75468 1 3 (HCC827) expression beadchip Head and neck cancer Affymetrix Human Genome U133 Cetuximab GSE21483 3 3 (SCC1) Plus 2.0 Array GEO, gene expression omnibus; GSE, gene expression series; S, sensitive; AR, acquired EGFR-TKI resistant; * Lung Cancer Cells Derived from Tumor Xenograft Model. Table S2. The performances of four penalized regression models. Model ACC precision recall F1 MCC AUROC BRIER Ridge 0.889 0.852 0.958 0.902 0.782 0.964 0.129 Lasso 0.944 0.957 0.938 0.947 0.889 0.991 0.042 Elastic Net 0.978 0.979 0.979 0.979 0.955 0.999 0.023 EPSGO Elastic Net 0.989 1.000 0.979 0.989 0.978 1.000 0.018 AUROC, area under curve of receiver operating characteristic; ACC, accuracy; MCC, Matthews correlation coefficient; EPSGO, Efficient Parameter Selection via Global Optimization algorithm. -
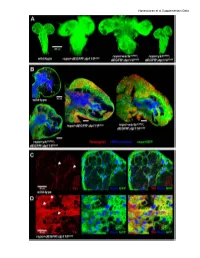
Supplementary Data Vigneswaran Et Al Supplementary Data
Vigneswaran et al Supplementary Data Vigneswaran et al Supplementary Data Figure S1: Yki is required for EGFR-PI3K-driven glial neoplasia in Drosophila (A) Optical projections of whole brain-nerve cord complexes from 3rd instar larvae approximately 130 hrs old. Dorsal view; anterior up. CD8-GFP (green) labels glial cell bodies. Compared to repo>dEGFRλ;dp110CAAX, warts knockdown (repo>wartsdsRNA; dEGFRλ;dp110CAAX) increased neoplastic brain overgrowth and yki knockdown (repo>ykidsRNA;dEGFRλ;dp110CAAX) decreased neoplastic brain overgrowth. (B) 3 µm optical projections of brain hemispheres, age-matched 3rd instar larvae. Frontal sections; anterior up; midline to left. Repo (red) labels glial cell nuclei; CD8-GFP (green) labels glial cell bodies; anti-HRP (blue) counter-stains for neurons and neuropil. (middle) repo>dEGFRλ;dp110CAAX showed increased glial cell numbers (red nuclei) compared to (upper left) wild-type. Compared to repo>dEGFRλ;dp110CAAX, (right) warts knockdown increased neoplastic glial cell numbers (red nuclei), whereas (lower left) yki knockdown reduced neoplastic glial cell numbers (red nuclei). (C, D) Low levels of Yki protein (red) was observed in wild-type central brain glia (white arrows, left panel in C) compared to high levels of cytoplasmic and nuclear Yki protein in dEGFRλ;dp110CAAX neoplastic glia (white arrows, left panel in D); Repo (blue) labels glial cell nuclei; CD8-GFP (green) labels glial cell bodies. Vigneswaran et al Supplementary Data Figure S2: YAP/TAZ expression confined to RTK-amplified tuMor cells and Maintained in patient-derived xenografts (A) On the left, immunohistochemical (IHC) staining in representative normal brain parenchyma in the cortex where YAP expression and TAZ expression was limited to vascular cells and was not detectable in normal neuronal and glial cells. -

Dependent Protein Kinase (PRKACA) Gene in Japanese Patients with Several Adrenal Adenomas Secreting Cortisol
Endocrine Journal 2014, 61 (8), 825-832 RAPID COMMUNICATION Somatic mutations of the catalytic subunit of cyclic AMP- dependent protein kinase (PRKACA) gene in Japanese patients with several adrenal adenomas secreting cortisol Yasuyo Nakajima1)*, Takashi Okamura1)*, Tamae Gohko1), Tetsurou Satoh1), Koshi Hashimoto1), Nobuyuki Shibusawa1), Atsushi Ozawa1), Sumiyasu Ishii1), Takuya Tomaru1), Kazuhiko Horiguchi1), Shuichi Okada1), Daisuke Takata2), Nana Rokutanda2), Jun Horiguchi2), Yoshito Tsushima3), Tetsunari Oyama4), Izumi Takeyoshi2) and Masanobu Yamada1) 1) Department of Medicine and Molecular Science, Gunma University Graduate School of Medicine, Maebashi 371-8511, Japan 2) Department of Thoracic and Visceral Organ Surgery, Gunma University Graduate School of Medicine, Maebashi 371-8511, Japan 3) Department of Diagnostic Radiology and Nuclear Medicine, Gunma University Graduate School of Medicine, Maebashi 371- 8511, Japan 4) Department of Diagnostic Pathology, Gunma University Graduate School of Medicine, Maebashi 371-8511, Japan Abstract. Somatic mutations of the catalytic subunit of the cyclic AMP-dependent protein kinase (PRKACA) gene have recently been identified in about 35% of cortisol-producing adenomas (CPAs), with the affected patients showing overt Cushing’s syndrome. Since we recently reported higher prevalence of mutations of the KCNJ5 gene and associations with autonomous cortisol secretion in Japanese aldosterone-producing adenomas than in Western countries, there might be different features of CPAs between Japan and the West. We therefore investigated mutations of the PRKACA gene in Japanese patients with several adrenal tumors secreting cortisol, including overt Cushing’s syndrome, subclinical Cushing’s syndrome, and aldosterone-producing adenomas (APAs) co-secreting cortisol operated on at Gunma University Hospital. Of the 13 patients with CPA who showed overt Cushing’s syndrome, 3 (23%) had recurrent somatic mutations of the PRKACA gene, p.L206R (c.617 T>G), and there were no mutations in subclinical Cushing’s syndrome. -

NIH Public Access Author Manuscript J Theor Biol
NIH Public Access Author Manuscript J Theor Biol. Author manuscript; available in PMC 2014 March 07. NIH-PA Author ManuscriptPublished NIH-PA Author Manuscript in final edited NIH-PA Author Manuscript form as: J Theor Biol. 2013 March 7; 320: . doi:10.1016/j.jtbi.2012.12.011. A predictive mathematical model of the DNA damage G2 checkpoint Kevin J. Kesselera, Michael L. Blinovb, Timothy C. Elstonc, William K. Kaufmanna, and Dennis A. Simpsona,* aDepartment of Pathology and Laboratory Medicine, Lineberger Comprehensive Cancer Center, Center for Environmental Health and Susceptibility, University of North Carolina at Chapel Hill, NC 27599-7255, USA bCenter for Cell Analysis and Modeling, University of Connecticut Health Center, 263 Farmington Avenue, Farmington, CT 06030-1507, USA cDepartment of Pharmacology, University of North Carolina at Chapel Hill,Chapel Hill, NC 27599-7260, USA Abstract A predictive mathematical model of the transition from the G2 phase in the cell cycle to mitosis (M) was constructed from the known interactions of the proteins that are thought to play significant roles in the G2 to M transition as well as the DNA damage- induced G2 checkpoint. The model simulates the accumulation of active cyclin B1/Cdk1 (MPF) complexes in the nucleus to activate mitosis, the inhibition of this process by DNA damage, and transport of component proteins between cytoplasm and nucleus. Interactions in the model are based on activities of individual phospho-epitopes and binding sites of proteins involved in G2/M. Because tracking phosphoforms leads to combinatorial explosion, we employ a rule-based approach using the BioNetGen software. The model was used to determine the effects of depletion or over-expression of selected proteins involved in the regulation of the G2 to M transition in the presence and absence of DNA damage.