Intraspecific Relationships and Variation of Two Lefua Species (Balitoridae, Cypriniformes) in the Tokai Region, Honshu, Japan
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Research on Prevention and Mitigation of Flood-Related Disasters in the World by Utilizing Integrated Risk Management Approach
-Priority Research Project- Research on prevention and mitigation of flood-related disasters in the world by utilizing integrated risk management approach Research Period: FY2005-2010 Project Leader: Director of Water-related Hazard Research Group TERAKAWA Akira Research Group: Water-related Hazard Research Group Cold-Region Hydraulic and Aquatic Environment Engineering Research Group Abstract: In order to make contribution for preventing and mitigating flood-related disasters in the world, this project covers several topics for integrated risk assessment and risk management in various natural and social conditions. The research topics in FY2006 include analysis of social vulnerability to flood disaster, flood forecasting and warning making use of satellite information, flood hazard mapping and experimental/analytical study on the behavior of tsunami wave running up into rivers. The outputs from the project are expected to be used as training materials for practical engineers of developing countries in charge of flood related disasters management. Key words: flood, tsunami, risk management, capacity building, forecasting, warning, hazard map -Individual Themes- A case study on asistance for strengthning flood damage mitigation measurers Budged: Grants for operating expenses General account Research Period: FY2006-2008 Research Team: Disaster Prevention Author: YOSHITANI Junichi TAKEMOTO Norimichi Abstract: This study is to summarize analyses of local vulnerability for water hazards and tangible measurers for strengthening damage mitigation systems on a regional basis. In fiscal 2006, we selected two countries of Philippines and Sri Lanka and did factor analysis of flood damages by means of literature research. Furthermore, regarding Bangladesh, we collected additional information mainly by hearings, formed a hypothesis and verified it by field survey. -

Representations of Pleasure and Worship in Sankei Mandara Talia J
Mapping Sacred Spaces: Representations of Pleasure and Worship in Sankei mandara Talia J. Andrei Submitted in partial fulfillment of the Requirements for the degree of Doctor of Philosophy in the Graduate School of Arts and Sciences Columbia University 2016 © 2016 Talia J.Andrei All rights reserved Abstract Mapping Sacred Spaces: Representations of Pleasure and Worship in Sankei Mandara Talia J. Andrei This dissertation examines the historical and artistic circumstances behind the emergence in late medieval Japan of a short-lived genre of painting referred to as sankei mandara (pilgrimage mandalas). The paintings are large-scale topographical depictions of sacred sites and served as promotional material for temples and shrines in need of financial support to encourage pilgrimage, offering travelers worldly and spiritual benefits while inspiring them to donate liberally. Itinerant monks and nuns used the mandara in recitation performances (etoki) to lead audiences on virtual pilgrimages, decoding the pictorial clues and touting the benefits of the site shown. Addressing themselves to the newly risen commoner class following the collapse of the aristocratic order, sankei mandara depict commoners in the role of patron and pilgrim, the first instance of them being portrayed this way, alongside warriors and aristocrats as they make their way to the sites, enjoying the local delights, and worship on the sacred grounds. Together with the novel subject material, a new artistic language was created— schematic, colorful and bold. We begin by locating sankei mandara’s artistic roots and influences and then proceed to investigate the individual mandara devoted to three sacred sites: Mt. Fuji, Kiyomizudera and Ise Shrine (a sacred mountain, temple and shrine, respectively). -
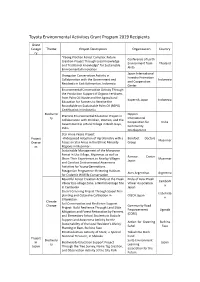
List of Previous Grant Projects
Toyota Environmental Activities Grant Program 2019 Recipients Grant Catego Theme Project Description Organization Country ry "Kaeng Krachan Forest Complex: Future Conference of Earth Creation Project Through Local Knowledge Environment from Thailand and Traditional Knowledge" for Sustainable Akita Environmental Innovation Japan International Orangutan Conservation Activity in Forestry Promotion Collaboration with the Government and Indonesia and Cooperation Residents in East Kalimantan, Indonesia Center Environmental Conservation Activity Through the Production Support of Organic Fertilizers from Palm Oil Waste and the Agricultural Kopernik Japan Indonesia Education for Farmers to Receive the Roundtable on Sustainable Palm Oil (RSPO) Certification in Indonesia Biodiversi Nippon Practical Environmental Education Project in ty International Collaboration with Children, Women, and the Cooperation for India Government in a Rural Village in Bodh Gaya, Community India Development Star Anise Peace Project Project -Widespread Adoption of Agroforestry with a Barefoot Doctors Myanmar Overse Focus on Star Anise in the Ethnic Minority Group as Regions in Myanmar- Sustainable Management of the Mangrove Forest in Uto Village, Myanmar, as well as Ramsar Center Share Their Experiences to Nearby Villages Myanmar Japan and Conduct Environmental Awareness Activities for Young Generations Patagonian Programme: Restoring Habitats Aves Argentinas Argentina for Endemic Wildlife Conservation Beautiful Forest Creation Activity at the Preah Pride of Asia: Preah -

Volki (Annelida: Clitellata: Propappidae) from Japan
JapaneseJapaneseSociety Society ofSystematicof Systematic Zoology Species Diversity, 2006, 11, 359-365 New Record of Propappus volki (Annelida: Clitellata: Propappidae) from Japan Takaaki Torii ll)Et`I CDnsuttants inc., 1334-5Riemon, Oigawa. Shida, Shizuoka, 421-CL212 .lapan E-mail: [email protected] (Received 1 November 2005; Accepted 11 July 2006) A species of freshwater oligochaete, PrQpoppus volki Michaelsen, 1916, is newly recorded from sand and gravel bottoms of several unpolluted streams in Honshu, Japan. The present material agrees well with the previous de- scriptiens of this species except that the spermathecal ampulla is shorter and the proboscis is more elomgate than those described earlier. This is the first record of the family Propappidae Coates, 1986 (New Japanese name: Ko- himemimizu-ka) and the genus Prql)qppus Miehaelsen, 1905 (New Japanese narne: Ko-himemimizu-zoku) in Japan. It provides evidence that the oligochaete fauna in Japan is more closely related to the Holarctic than to the Sino-Indian zoogeographical region, PrQpqppus votici can be regarded as a good bio-indicator fbr unpolluted water. Key Words: oligochaetes, Clitellata, Propappidae, Propappus volki, new record, Japan. Introduction The genus PrQpqppus was erected by Michaelsen (1905) fbr PrQpappus gtandu- losus Michaelsen, 1905 in the family Enchytraeidae. Afterwards Coates (1986) cre- ated the family Propappidae for the single genus PrQpappus on the basis of a unique combination of setal and genital characteristics distinctly different from those ef the Enchytraeidae. Only three species of Propampus have so far been described from freshwater habitats of the Palaearctic region, Among them, Prqpqppus glandulosus has been recorded from Lake Baikal and the Angara and Yenisey Rivers in central Russia (Michaelsen 1905; Coates 1986), PrQpqppus arhynchotus Sokolskaja, 1972 has been recorded only from the Kamchatka Peninsula and Amur basin in the Russian Far East (Timm 1994, 1999a). -

Flood Loss Model Model
GIROJ FloodGIROJ Loss Flood Loss Model Model General Insurance Rating Organization of Japan 2 Overview of Our Flood Loss Model GIROJ flood loss model includes three sub-models. Floods Modelling Estimate the loss using a flood simulation for calculating Riverine flooding*1 flooded areas and flood levels Less frequent (River Flood Engineering Model) and large- scale disasters Estimate the loss using a storm surge flood simulation for Storm surge*2 calculating flooded areas and flood levels (Storm Surge Flood Engineering Model) Estimate the loss using a statistical method for estimating the Ordinarily Other precipitation probability distribution of the number of affected buildings and occurring disasters related events loss ratio (Statistical Flood Model) *1 Floods that occur when water overflows a river bank or a river bank is breached. *2 Floods that occur when water overflows a bank or a bank is breached due to an approaching typhoon or large low-pressure system and a resulting rise in sea level in coastal region. 3 Overview of River Flood Engineering Model 1. Estimate Flooded Areas and Flood Levels Set rainfall data Flood simulation Calculate flooded areas and flood levels 2. Estimate Losses Calculate the loss ratio for each district per town Estimate losses 4 River Flood Engineering Model: Estimate targets Estimate targets are 109 Class A rivers. 【Hokkaido region】 Teshio River, Shokotsu River, Yubetsu River, Tokoro River, 【Hokuriku region】 Abashiri River, Rumoi River, Arakawa River, Agano River, Ishikari River, Shiribetsu River, Shinano -

How Is the Gap Between the Concept and Practice of Integrated Sediment Management Bridged?
K-3 Fourth International Conference on Scour and Erosion 2008 HOW IS THE GAP BETWEEN THE CONCEPT AND PRACTICE OF INTEGRATED SEDIMENT MANAGEMENT BRIDGED? Koh-ichi FUJITA Member of JSCE, Research Coordinator for Environmental Affairs, Environment Dept., National Institute for Land and Infrastructure Management (NILIM), Ministry of Land, Infrastructure, Transport and Tourism (MLIT) (Asahi 1, Tsukuba-shi, Ibaraki-ken, 305-0804, Japan) E-mail: [email protected] In Japan, various and intensive modifications to river systems through projects for sediment/flood control, water resources development, electric power supply, river improvement and so on have played extremely important roles in mitigating flood/sediment related disasters and improving our lives and society. At the same time, they have changed river-basin-scale sediment transport systems, bringing new problems with their system soundness in terms of continuity, sustainability and ecological functions. Perceiving that this shows limitations of a locally optimized approach by the area and purpose, which had been taken because of its efficiency, the government has already laid out the concept of “integrated management of a sediment transport system” as a new national policy direction. However, there still appears to be the gap between the concept and its practice, which may retard sweeping development of the new policy. Instead of seeking for “magic technology” that alone can bridge the gap, this overview paper stresses three keys to firmly establishing integrated sediment management: (a) grasping and sharing an overall image of a sediment transport system by using a “common language” that macroscopically describes sediment transport phenomena, not being limited to excessively precise analysis; (b) appropriately performing a diagnosis to identify the structure of problems through a scenario-driven approach, not being preoccupied by stereotyped thoughts; (c) prioritizing the development of component technologies and linking them with policy setting & implementation processes. -

Vulnerability to Flood Risks in Japanese Urban Areas: Crisis Management and Emergency Response for Efficient Evacuation Management
Flood Recovery, Innovation and Reponse IV 61 Vulnerability to flood risks in Japanese urban areas: crisis management and emergency response for efficient evacuation management M. Thomas & T. Tsujimoto Nagoya University, Department of Civil Engineering, Japan Abstract Today, flood risk in Japan occurs mainly in high density populated areas, as a consequence of the rapid urban development of the deltaic plains of Japan during the second half of the 20th century. At the end of the 20th century risk management began to shift from mainly structural management to a more “integrated” management. The evacuation process is one of the factors revealing this shift. In Nagoya the evacuation process enhancement started with the Tokai flood disaster (September 2000) and continues to this day. The most recent flood events (urban flood of 2008 and typhoon No. 14 of 2011) highlight, however, how the crisis management can still be vulnerable regarding evacuation. Our research intends to assess the vulnerability factors of the crisis management system, and especially of the evacuation process through interviews and a questionnaire analysis method, in order to propose an integrated way of dealing with evacuation in the case of a flood, imputing on GIS geographical as well as social characteristics and evacuation patterns. Our research shows that the evacuation process is effective despite low evacuation rate during past flood event. In that regard improving the evacuation process cannot be separated from the improvement of informational tools, but it can be seen that the possession of hazard maps have few impact on evacuation decision. The efficiency of the evacuation process in the case of a small to moderate flood event could therefore be enhanced as the large-scale evacuation broadcast tends to target a population in which more than half of the people do not need to evacuate. -

Note Sensory Analysis of the Taste of Ayu Sampled from Different River
Food Sci. Technol. Res., 19 (3), 479–483, 2013 Note Sensory Analysis of the Taste of Ayu Sampled from Different River Environments 1* 2 1 1 Noriko HattoRi , Asako uCHida , Mitsuaki SaNo and Tetuo MuRakaMi 1 Graduate School of Human Life Science, Nagoya Women’s University, 3-40 Shioji-cho, Mizuho-ku, Nagoya 467-8610, Japan 2 Toyota Yahagi River Institute, 2-19 Nishi-machi, Toyota 471-0025, Japan Received December 18, 2012; Accepted January 22, 2013 It is widely believed that attached diatoms are the best food source for freshwater ayu Plecoglossus altivelis (Temminck et Schlegel) in Japan. However, sensory testing by a panel consisting of 40 untrained tasters and analyses of gut content showed that there was no significant correlation between preference scores for fish taste and the dominance of diatoms in the algal assemblages fed upon by ayu in the Yahagi River, Aichi Prefecture, Central Japan. Ayu that fed on the filamentous blue-green algae Homeothrix varians was evaluated higher for aroma and fat content than those that fed on diatoms. Ayu feeding on diatoms were collected in the lower reaches of the Yahagi River which receive domestic sewage. In the ab- sence of more detailed analyses, exposure to sewage is considered to confer a characteristic aroma on Ayu. Keywords: attached algae, Ayu, gut content, sensory testing, Yahagi River Introduction also doubtful. For example, Hodoki et al. (2003) showed that Ayu, Plecoglossus altivelis (Temminck et Schlegel), a there was no significant difference in body length and weight member of Plecoglossidae, is one of the most popular fresh- between two ayu populations that fed on either diatoms or water fishes in Japan. -

Japanese Experience on Structural Measures for Flood Management
Japanese experience on Structural Measures for Flood Management Kazuhiko FUKAMI Hydrologic Engineering Research Team, Public Works Research Institute (PWRI), Japan Kenji KANAO and Katsuhisa SHIOJI River Bureau, Ministry of Land, Infrastructure and Transport (MLIT), Japan Rivers in Japan are steep. Rivers in Japan tend to be steep, short and rapid flowing. Rhine River Loire River Joganji River Colorado River Abe River Shinano River Tone River Chikugo River Seine River Yoshino River Kitakami River Mekong River Elevation River mouth Distance from river mouth (km) Comparison of the longitudinal profiles of rivers in Japan and other countries -1- Fifty percent of population and 75% of property are concentrated in floodplains accounting for only 10% of total land area. Alluvial plains Other areas (areas lower than river stage in times of flood) Property Population Land area -2- Land use changes in the left-bank area of the Ara River Floodway in the past 100 years 明治188215年 Present現在 Adachi Ward Adachi足立区 Ward Katsushika Katsushika葛飾区 Ward Ward Edogawa Edogawa江戸川区 Ward Ward -3- Major storm and flood disaster after WWII ~ Typhoon Kathleen (September, 1947) ~ Number of persons killed: 1077 Number of persons missing: 853 Number of persons injured: 1,547 Number of houses completely or partially destroyed: 9,298 Above-floor-level/below-floor-level inundation: 384,743 Katsuhika Ward, Tokyo Areas inundated by the September 1947 flood Failure of the levee along the Tone River in the Tone River System (134.5km from river mouth) -4- Changes in the number of persons killed by storms and floods Disaster o Nishi Nihon T I Kano I Disaster o T Nishi NihonHeav Second M Nag Sanin T T M Ky Har Rain Fukushim Rain Disaster Hiroshi Tokai He s s o ah e y y y t y phoon No. -

Dam, Construction, Red Tide, Surface Area, Bay, Estuary
World Environment 2012, 2(6): 120-126 DOI: 10.5923/j.env.20120206.03 Relationship between Red Tide Occurrences in Four Japanese Bays and Dam Construction Kunio Ueda Department of Biological Resources Management, School of Environmental Science, The University of Shiga Prefecture, Japan Abstract Since about half a century ago, red tide has been occurring in many coastal places of Japan, such as Tokyo Bay, Ise Bay, Osaka Bay, and Ariake Sea. Red tide is algal accumulation that could be a result of eutrophication in bays and lakes. At the same time, dams have been constructed in Japan on rivers that flow into the bays where red tide has been occurring. The correlation between red tide occurrence and dam construction in Japan was researched using the data of many government organizations. The results indicate that the construction of dams influences the occurrences of red tide. When a dam is built on a river, there is a tendency for red tide to result in an estuary of that river a few years later. The number of red tide occurrences is related to the surface area of the dam: as the surface area of a constructed dam increases, the number of red tide occurrences in a bay increases. Thus, the construction of dams seems to cause eutrophication in bays and lakes. Because it seemed that small particles flowed from dams contain nutrients that stimulate the growth of algae. Ke ywo rds Red Tide, Dam, Construction, Coastal Area, Surface Area, Es tuary In these cases, other factors should be discussed for 1. Introduction solving the problem of red tide occurrence. -

(Watershed) ESD Model in Chubu Area, Aichi-Nagoya, Japan
The Bioregional (Watershed) ESD Model in Chubu Area, Aichi-Nagoya, Japan Reita Furusawa - Associate Professor, Chubu Ins8tute of Advanced Studies/Interna8onal ESD Center, Chubu University - Secretary General, RCE Chubu ~ RCEs around the World ~ RCEs in Japan RCE G.Sendai RCE Yokohama RCE Chubu RCE Hyogo Kobe RCE Okayama RCE Kitakyushu RCE Chubu Network Universities Administrative Institutions Kachigawa-Ekimae Shopping Street Aichi Gakuin University Aichi Prefectural Government Promotion Association Aichi Prefectural University Gifu Prefectural Government Citizens Environmental Foundation Chubu University Mie Prefecture Gifu NPO Center Nagoya Institute of Technology Kasugai City Gifu Industrial Hemp Association Nagoya City University Nagoya City Hall International Green Citizen Association Nagoya University Chubu Regional Environment Office (IGCA) Gifu University Chubu Bureau of Economy, Trade and Industry METI Alaska, its spirits and Michio Hoshino Nihon Fukushi University "Chubu District Transport Bureau ,Ministry of Land, Support Center for Sustainable Regional Meijo University Infrastructure, Transport And Tourism " Design Mie University "Chubu Regional Bureau, Ministry of Land, DEE21(Digital Economy & Enterprise for Infrastructure, Transport And Tourism " the 21st. Century) Tokai Regional Agricultural Administration Office, Nagoya International Center High and Junior High School Ministry of Agriculture, Forestry and Fisheries Nagoya UNESCO Chubu University Daiichi High School Environmental Partnership Office Chubu Nissin Nature Observation -

Directorate General of Water Resources Ministry of Public Works Republic of Indonesia 1 • Information for Preventive Investment
Water –related Disaster Risk Information for Risk Reduction June 23, 2014 The 6th Asian Ministerial Conference on Disaster Risk reduction in Bangkok, Thailand Directorate General of Water Resources Ministry of Public Works Republic of Indonesia 1 • Information for Preventive Investment • Information for Preparedness and Response • Summary 2 Disaster Risk Management • Mitigation DISASTER Response to prevent disasters Preparedness • Preparedness to ensure effective response Recovery • Response Mitigation to reduce adverse impacts during the flooding • Recovery to assist the affected communities to rebuild themselves We need to consider what information is necessary in accordance with each stage 3 Effect of Preventive Investment • Investment for prevention can reduce disaster damages largely In case of “TOKAI devastating flood” in Japan 2000, Prior investments of JPY71.6 billion made it possible to reduce disaster damage by about JPY550 billion. Effect of disaster prevention projects: Equivalent of approx. Amount of damage: JPY550 billion Approx. Waterlogged conditions due to heavy downpours in Tokai region JPY670 billion After implementation of projects: Estimated amount of Projects costs: damage:Approx. * Special Emergency Project for Shonai River JPY 120 billion JPY 71.6 billion in total and Shin River Disaster Prevention Strategies(2000 to 2004). Amount of disaster damage Estimated amount of Costs for preventing incurred from heavy damage after recurrence of disasters* downpour in Tokai region implementation of projects 4 Number of Fatalities