Living Between Rapids: Genetic Structure and Hybridization in Botos (Cetacea: Iniidae: Inia Spp.) of the Madeira River, Brazil
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Inia Araguaiaensisand Inia Geoffrensis
Interactions between children, teenagers and botos (Inia araguaiaensis and Inia geoffrensis) in markets and fairs of Eastern Amazon Angélica Lúcia Figueiredo Rodrigues, Gabriel Melo-Santos, Iara Ramos-Santos, Ana Marta Andrade, Danilo Leal Arcoverde, Leonardo Sena, Maria Luisa da Silva Date of deposit 09 05 2019 Document version Author’s accepted manuscript Access rights © 2019, Elsevier Ltd. All rights reserved. This work is made available online in accordance with the publisher’s policies. This is the author created, accepted version manuscript following peer review and may differ slightly from the final published version. Citation for Figueiredo Rodrigues, A.L., Melos-Santos, G., and Ramos-Santos, published version I. (2019). Interactions between children, teenagers and botos (Inia araguaiaensis and Inia geoffrensis) in markets and fairs of Eastern Amazon. Ocean and Coastal Management, 172: pp137- 145. Link to published https://doi.org/j.ocecoaman.2019.02.005 version Full metadata for this item is available in St Andrews Research Repository at: https://research-repository.st-andrews.ac.uk/ 1 Abstract In this study we present the first record of interactions (ludic behavior, swimming and induced feeding) involving river dolphins known as botos (Inia sp.) and children/adolescents (from eight to 17 years old) at markets and fairs at the river front of two localities of Pará State, Brazil. We registered the reports of the participants using semi-structured interviews while they were working at the markets or when they were in the water to interact with river dolphins. We registered dolphins and human behavior using the ad libitum method. Most of the children interacting with botos were boys - we observed only two girls swimming with the dolphins. -

Encyclopedia of Marine Mammals, Second Edition
1188 Tucuxi and Guiana Dolphin continues to grow and in the United States, public support stands Chance , P. ( 1994 ). “ Learning and Behavior , ” 3rd Ed. Brooks/Cole fi rmly behind both the MMPA and marine mammal facilities. More Publishing Company , Belmont . people are now enjoying the benefi ts of new and exciting training Cole , K. C. , Van Tilburg , D. , BurchVernon , A. , and Riccio , D. C. ( 1996). programs, shows, presentations, interaction opportunities, and scien- The importance of context in the US preexposure effect in CTA: Novel tifi c discoveries, all facilitated through behavior management. versus latently inhibited contextual stimuli . Lear. Motiv. 27 , 362 – 374 . Domjan , M. ( 1993 ). “ The Principles of Learning and Behavior , ” 3rd Ed. By maintaining a healthy captive population of various marine Brooks/Cole Publishing Company , Belmont . mammal species, comparative data are generated to assist in under- Honig , W. K. , and Staddon , J. E. R. ( 1977 ). “ The Handbook of Operant standing wild animals, and these facilities continue to give material Behavior . ” Prentice-Hall, Inc , Englewood Cliffs . support to important research and conservation initiatives. In addi- Kazdin , A. E. ( 1994 ). “ Behavior Modifi cation in Applied Settings , ” 5th tion, these facilities act as part of the Marine Mammal Stranding Ed. Brooks/Cole Publishing Company , Belmont . Network, assisting NOAA/NMFS in the rescue, housing, and care Marine Mammal Permits and Authorizations. (2006). [Accessed online of stranded wild animals where expertise in medical care can be July 5, 2007]. Available from World Wide Web: http://www.nmfs. applied. These facilities also develop animal management and hus- noaa.gov/pr/permits/mmpa_permits.htm bandry skills in staff members who are also able to assist in health Marine Mammal Poll. -

Cetaceans: Whales and Dolphins
CETACEANS: WHALES AND DOLPHINS By Anna Plattner Objective Students will explore the natural history of whales and dolphins around the world. Content will be focused on how whales and dolphins are adapted to the marine environment, the differences between toothed and baleen whales, and how whales and dolphins communicate and find food. Characteristics of specific species of whales will be presented throughout the guide. What is a cetacean? A cetacean is any marine mammal in the order Cetaceae. These animals live their entire lives in water and include whales, dolphins, and porpoises. There are 81 known species of whales, dolphins, and porpoises. The two suborders of cetaceans are mysticetes (baleen whales) and odontocetes (toothed whales). Cetaceans are mammals, thus they are warm blooded, give live birth, have hair when they are born (most lose their hair soon after), and nurse their young. How are cetaceans adapted to the marine environment? Cetaceans have developed many traits that allow them to thrive in the marine environment. They have streamlined bodies that glide easily through the water and help them conserve energy while they swim. Cetaceans breathe through a blowhole, located on the top of their head. This allows them to float at the surface of the water and easily exhale and inhale. Cetaceans also have a thick layer of fat tissue called blubber that insulates their internals organs and muscles. The limbs of cetaceans have also been modified for swimming. A cetacean has a powerful tailfin called a fluke and forelimbs called flippers that help them steer through the water. Most cetaceans also have a dorsal fin that helps them stabilize while swimming. -

Food Consumption and Body Measurements of Amazon River Dolphins (Inia Geoffrensis)
Aquatic Mammals 1999, 25.3, 173–182 Food consumption and body measurements of Amazon river dolphins (Inia geoffrensis) R. A. Kastelein1, B. Neurohr2, S. H. Nieuwstraten1 and P. R. Wiepkema3 1Harderwijk Marine Mammal Park, Strandboulevard Oost 1, 3841 AB Harderwijk, The Netherlands 2Tiergarten Nu¨rnberg, Am Tiergarten 30, D90480 Nu¨rnberg, Germany 3Emeritus Professor of Ethology, Wageningen Agricultural University, Stationsweg 1, 6861 EA Oosterbeek, The Netherlands Abstract dolphin is the most widespread freshwater dolphin in the world, its distribution is limited compared to This report is on the food consumption of 3 male that of most marine odontocetes and it is therefore Amazon river dolphins which were housed in water very likely to become a threatened species. For the of between 27 and 29 C at Duisburg Zoo, management of Amazon river dolphins in the wild, Germany. The food consumption of 2 animals was information is needed about the population size, recorded for 17 successive complete calendar years, age composition and sex ratio, seasonal distribu- that of the third animal for 3 complete successive tion, diet, energy requirements relative to seasonal years. In male 002 the total annual food intake prey distribution and density, and about competi- increased to 1280 kg at the age of 10 years, after tion with other animals and with fisheries. This which it decreased slightly and stabilised at around information could facilitate prey management 1170 kg/year. Male 001 was adult on arrival at the to allow for a certain number of Amazon river zoo. His annual food intake fluctuated at around dolphins in their distribution area. -
Global Patterns in Marine Mammal Distributions
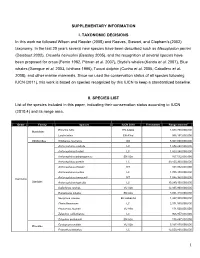
SUPPLEMENTARY INFORMATION I. TAXONOMIC DECISIONS In this work we followed Wilson and Reeder (2005) and Reeves, Stewart, and Clapham’s (2002) taxonomy. In the last 20 years several new species have been described such as Mesoplodon perrini (Dalebout 2002), Orcaella heinsohni (Beasley 2005), and the recognition of several species have been proposed for orcas (Perrin 1982, Pitman et al. 2007), Bryde's whales (Kanda et al. 2007), Blue whales (Garrigue et al. 2003, Ichihara 1996), Tucuxi dolphin (Cunha et al. 2005, Caballero et al. 2008), and other marine mammals. Since we used the conservation status of all species following IUCN (2011), this work is based on species recognized by this IUCN to keep a standardized baseline. II. SPECIES LIST List of the species included in this paper, indicating their conservation status according to IUCN (2010.4) and its range area. Order Family Species IUCN 2010 Freshwater Range area km2 Enhydra lutris EN A2abe 1,084,750,000,000 Mustelidae Lontra felina EN A3cd 996,197,000,000 Odobenidae Odobenus rosmarus DD 5,367,060,000,000 Arctocephalus australis LC 1,674,290,000,000 Arctocephalus forsteri LC 1,823,240,000,000 Arctocephalus galapagoensis EN A2a 167,512,000,000 Arctocephalus gazella LC 39,155,300,000,000 Arctocephalus philippii NT 163,932,000,000 Arctocephalus pusillus LC 1,705,430,000,000 Arctocephalus townsendi NT 1,045,950,000,000 Carnivora Otariidae Arctocephalus tropicalis LC 39,249,100,000,000 Callorhinus ursinus VU A2b 12,935,900,000,000 Eumetopias jubatus EN A2a 3,051,310,000,000 Neophoca cinerea -

Riverine and Marine Ecotypes of Sotalia Dolphins Are Different Species
Marine Biology (2005) 148: 449–457 DOI 10.1007/s00227-005-0078-2 RESEARCH ARTICLE H.A. Cunha Æ V.M.F. da Silva Æ J. Lailson-Brito Jr M.C.O. Santos Æ P.A.C. Flores Æ A.R. Martin A.F. Azevedo Æ A.B.L. Fragoso Æ R.C. Zanelatto A.M. Sole´-Cava Riverine and marine ecotypes of Sotalia dolphins are different species Received: 24 December 2004 / Accepted: 14 June 2005 / Published online: 6 September 2005 Ó Springer-Verlag 2005 Abstract The current taxonomic status of Sotalia species cific status of S. fluviatilis ecotypes and their population is uncertain. The genus once comprised five species, but structure along the Brazilian coast. Nested-clade (NCA), in the twentieth century they were grouped into two phylogenetic analyses and analysis of molecular variance (riverine Sotalia fluviatilis and marine Sotalia guianensis) of control region sequences showed that marine and that later were further lumped into a single species riverine ecotypes form very divergent monophyletic (S. fluviatilis), with marine and riverine ecotypes. This groups (2.5% sequence divergence; 75% of total molec- uncertainty hampers the assessment of potential impacts ular variance found between them), which have been on populations and the design of effective conservation evolving independently since an old allopatric fragmen- measures. We used mitochondrial DNA control region tation event. This result is also corroborated by cyto- and cytochrome b sequence data to investigate the spe- chrome b sequence data, for which marine and riverine specimens are fixed for haplotypes that differ by 28 (out Communicated by J. P. -

Translocation of Trapped Bolivian River Dolphins (Inia Boliviensis)
J. CETACEAN RES. MANAGE. 21: 17–23, 2020 17 Translocation of trapped Bolivian river dolphins (Inia boliviensis) ENZO ALIAGA-ROSSEL1,3AND MARIANA ESCOBAR-WW2 Contact e-mail: [email protected] ABSTRACT The Bolivian river dolphin, locally known as the bufeo, is the only cetacean in land-locked Bolivia. Knowledge about its conservation status and vulnerability to anthropogenic actions is extremely deficient. We report on the rescue and translocation of 26 Bolivian river dolphins trapped in a shrinking segment of the Pailas River, Santa Cruz, Bolivia. Several institutions, authorities and volunteers collaborated to translocate the dolphins, which included calves, juveniles, and pregnant females. The dolphins were successfully released into the Río Grande. Each dolphin was accompanied by biologists who assured their welfare. No detectable injuries occurred and none of the dolphins died during this process. If habitat degradation continues, it is likely that events in which river dolphins become trapped in South America may happen more frequently in the future. KEYWORDS: BOLIVIAN RIVER DOLPHIN; HABITAT DEGRADATION; CONSERVATION; STRANDINGS; TRANSLOCATION; SOUTH AMERICA INTRODUCTION distinct species, geographically isolated from the boto or Small cetaceans are facing several threats from direct or Amazon River dolphin (I. geoffrensis) (Gravena et al., 2014; indirect human impacts (Reeves et al., 2000). The pressure Ruiz-Garcia et al., 2008). This species has been categorised on South American and Asian river dolphins is increasing; by the Red Book of Wildlife Vertebrates of Bolivia as evidently, different river systems have very different problems. Vulnerable (VU), highlighting the need to conserve and Habitat degradation, dam construction, modification of river protect them from existing threats (Aguirre et al., 2009). -

Full Text in Pdf Format
Vol. 45: 269–282, 2021 ENDANGERED SPECIES RESEARCH Published July 29 https://doi.org/10.3354/esr01133 Endang Species Res OPEN ACCESS Home range and movements of Amazon river dolphins Inia geoffrensis in the Amazon and Orinoco river basins Federico Mosquera-Guerra1,2,*, Fernando Trujillo1, Marcelo Oliveira-da-Costa3, Miriam Marmontel4, Paul André Van Damme5, Nicole Franco1, Leslie Córdova5, Elizabeth Campbell6,7,8, Joanna Alfaro-Shigueto6,7,8, José Luis Mena9, Jeffrey C. Mangel6,7,8, José Saulo Usma Oviedo3, Juan D. Carvajal-Castro10,11, Hugo Mantilla-Meluk12,13, Dolors Armenteras-Pascual2 1Fundación Omacha, 111211 Bogotá, D.C., Colombia 2Grupo de Ecología del Paisaje y Modelación de Ecosistemas-ECOLMOD, Departamento de Biología, Universidad Nacional de Colombia, 111321 Bogotá, D.C., Colombia 3World Wildlife Fund (WWF) − Brazil, Colombia, and Peru, Rue Mauverney 28, 1196 Gland, Switzerland 4Instituto de Desenvolvimento Sustentável Mamirauá, 69.553-225 Tefé (AM), Brazil 5Faunagua, 31001 Sacaba-Cochabamba, Bolivia 6ProDelphinus, 15074 Lima, Peru 7School of BioSciences, University of Exeter, Penryn, Cornwall TR10 9EZ, UK 8Carrera de Biología Marina, Universidad Cientifíca del Sur, 15067 Lima, Peru 9Museo de Historia Natural Vera Alleman Haeghebaert, Universidad Ricardo Palma, 1801 Lima, Peru 10Grupo de Investigación en Evolución, Ecología y Conservación (EECO), Programa de Biología, Universidad del Quindío, 630004 Armenia, Colombia 11Department of Biological Sciences, St. John’s University, 11366 Queens, NY, USA 12Grupo de Investigación en Desarrollo y Estudio del Recurso Hídrico y el Ambiente (CIDERA), Programa de Biología, Universidad del Quindío, 630004 Armenia, Colombia 13Centro de Estudios de Alta Montaña, Universidad del Quindío, 630004 Armenia, Colombia ABSTRACT: Studying the variables that describe the spatial ecology of threatened species allows us to identify and prioritize areas that are critical for species conservation. -

Phylogeny of All Major Groups of Cetaceans Based on DNA Sequences from Three Mitochondrial Genes
Phylogeny of All Major Groups of Cetaceans Based on DNA Sequences from Three Mitochondrial Genes Michel C. Milinkovitch,* Axe1 Meyer) and Jeffrey R. Powell * *Department of Biology, Yale University and +Department of Ecology and Evolution, State University of New York at Stony Brook Traditionally, living cetaceans (order Cetacea) are classified into two highly distinct suborders: the echolocating toothed whales, Odontoceti, and the filter-feeding baleen whales, Mysticeti. A molecular phylogeny based on 1,352 base pairs of two mitochondrial ribosomal gene segments and the mitochondrial cytochrome b gene for all major groups of cetaceans contradicts this long-accepted taxonomic subdivision. One group of toothed whales, the sperm whales, is more closely related to the morphologically highly divergent baleen whales than to other odontocetes. This finding suggests that the suborder Odontoceti constitutes an unnatural grouping and challenges the conventional scenario of a long, independent evolutionary history of odontocetes and mysticetes. The superfamily Delphinoidea (dolphins, porpoises, and white whales) appears to be monophyletic; the Amazon River dolphin, Inia geofiensis, is its sister species. This river dolphin is genetically more divergent from the morphologically similar marine dolphins than the sperm whales are from the morphologically dissimilar baleen whales. The phylogenetic relationships among the three families of Delphinoidea remain uncertain, and we suggest that the two cladogenetic events that generated these three clades occurred within a very short period of time. Among the baleen whales, the bowhead is basal, and the gray whale is the sister species to the rorquals (family Balaenopteridae). The phylogenetic position of beaked whales (Ziphioidea) remains weakly supported by molecular data. -

Marine Mammal Taxonomy
Marine Mammal Taxonomy Kingdom: Animalia (Animals) Phylum: Chordata (Animals with notochords) Subphylum: Vertebrata (Vertebrates) Class: Mammalia (Mammals) Order: Cetacea (Cetaceans) Suborder: Mysticeti (Baleen Whales) Family: Balaenidae (Right Whales) Balaena mysticetus Bowhead whale Eubalaena australis Southern right whale Eubalaena glacialis North Atlantic right whale Eubalaena japonica North Pacific right whale Family: Neobalaenidae (Pygmy Right Whale) Caperea marginata Pygmy right whale Family: Eschrichtiidae (Grey Whale) Eschrichtius robustus Grey whale Family: Balaenopteridae (Rorquals) Balaenoptera acutorostrata Minke whale Balaenoptera bonaerensis Arctic Minke whale Balaenoptera borealis Sei whale Balaenoptera edeni Byrde’s whale Balaenoptera musculus Blue whale Balaenoptera physalus Fin whale Megaptera novaeangliae Humpback whale Order: Cetacea (Cetaceans) Suborder: Odontoceti (Toothed Whales) Family: Physeteridae (Sperm Whale) Physeter macrocephalus Sperm whale Family: Kogiidae (Pygmy and Dwarf Sperm Whales) Kogia breviceps Pygmy sperm whale Kogia sima Dwarf sperm whale DOLPHIN R ESEARCH C ENTER , 58901 Overseas Hwy, Grassy Key, FL 33050 (305) 289 -1121 www.dolphins.org Family: Platanistidae (South Asian River Dolphin) Platanista gangetica gangetica South Asian river dolphin (also known as Ganges and Indus river dolphins) Family: Iniidae (Amazon River Dolphin) Inia geoffrensis Amazon river dolphin (boto) Family: Lipotidae (Chinese River Dolphin) Lipotes vexillifer Chinese river dolphin (baiji) Family: Pontoporiidae (Franciscana) -

Neophocaena Phocaenoides (Cuvier, 1829) PHOCO Neoph 1 PFI
click for previous page 192 Marine Mammals of the World Neophocaena phocaenoides (Cuvier, 1829) PHOCO Neoph 1 PFI FAO Names: En - Finless porpoise; Fr - Marsouin aptère; Sp - Marsopa lisa o sin aleta. Fig. 382 Neophocaena phocaenoides Distinctive Characteristics: As the name im- plies, finless porpoises have no dorsal fin, and this is their most distinctive characteristic. In some ways, they resemble small, slender white whales. The head is beakless; the rounded forehead rises steeply from the snout tip. The body shape, in general, is more slender than in other porpoises. The finless porpoise is soft and mushy, and the neck is very flexible. Instead of a dorsal fin, the finless porpoise has an area of small bumps or DORSAL VIEW tubercles on its back, running from just forward of midback to the tail stock. The trailing edge of the flukes is concave and the flippers are large, ending in rounded tips. Regional differences in body size and morphology have been documented, with Yangtze River animals apparently representing a separate stock. The common name that was used in the past, “finless black porpoise,” apparently resulted from descriptions of dead animals, after post-mortem VENTRAL VIEW darkening. In most areas, finless porpoises are grey in colour, with lighter areas on the throat and around the genitals. Older animals are generally lighter grey than juveniles. In the Yangtze River population, they are very dark grey, nearly black. Tooth counts range from 13 to 22 in each tooth row. DORSAL VIEW OF MANDIBLE LATERAL VIEW Fig. 383 Skull Cetacea - Odontoceti - Phocoenidae 193 Can be confused with: The smooth back of the finless porpoise should make it easy to distinguish from other species, such as the lrrawaddy dolphin (p. -

Review of Small Cetaceans. Distribution, Behaviour, Migration and Threats
Review of Small Cetaceans Distribution, Behaviour, Migration and Threats by Boris M. Culik Illustrations by Maurizio Wurtz, Artescienza Marine Mammal Action Plan / Regional Seas Reports and Studies no. 177 Published by United Nations Environment Programme (UNEP) and the Secretariat of the Convention on the Conservation of Migratory Species of Wild Animals (CMS). Review of Small Cetaceans. Distribution, Behaviour, Migration and Threats. 2004. Compiled for CMS by Boris M. Culik. Illustrations by Maurizio Wurtz, Artescienza. UNEP / CMS Secretariat, Bonn, Germany. 343 pages. Marine Mammal Action Plan / Regional Seas Reports and Studies no. 177 Produced by CMS Secretariat, Bonn, Germany in collaboration with UNEP Coordination team Marco Barbieri, Veronika Lenarz, Laura Meszaros, Hanneke Van Lavieren Editing Rüdiger Strempel Design Karina Waedt The author Boris M. Culik is associate Professor The drawings stem from Prof. Maurizio of Marine Zoology at the Leibnitz Institute of Wurtz, Dept. of Biology at Genova Univer- Marine Sciences at Kiel University (IFM-GEOMAR) sity and illustrator/artist at Artescienza. and works free-lance as a marine biologist. Contact address: Contact address: Prof. Dr. Boris Culik Prof. Maurizio Wurtz F3: Forschung / Fakten / Fantasie Dept. of Biology, Genova University Am Reff 1 Viale Benedetto XV, 5 24226 Heikendorf, Germany 16132 Genova, Italy Email: [email protected] Email: [email protected] www.fh3.de www.artescienza.org © 2004 United Nations Environment Programme (UNEP) / Convention on Migratory Species (CMS). This publication may be reproduced in whole or in part and in any form for educational or non-profit purposes without special permission from the copyright holder, provided acknowledgement of the source is made.