The Monophyly of Toothed Whales and the Paraphyly of River Dolphins
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Distribution of the Atlantic Bottlenose Dolphin (Tursiops Truncatus) in the Chesapeake Bay Drainage in Virginia
Banisteria, Number 11, 1998 33 © 1998 by the Virginia Natural History Society Distribution of the Atlantic Bottlenose Dolphin (Tursiops truncatus) in the Chesapeake Bay Drainage in Virginia Thomas F. Wilcox Virginia Department of Game and Inland Fisheries 4010 West Broad Street Richmond VA 23230 Susan G. Barco & W. Mark Swingle Virginia Marine Science Museum Stranding Program 717 General Booth Boulevard Virginia Beach, VA 23451 INTRODUCTION the Virginia Department of Environmental Quality, was completed by federal and state agencies, members of Coastal and estuarine Atlantic ,bottlenose dolphins academia, and conservation organizations. The overall (Tursiops truncatus) in Virginia are part of the coastal objective of the plan is to protect, manage, and enhance migratory stock listed as depleted under the Marine marine mammal populations and promote education, Mammal Protection Act (Wang et al., 1994). Live participation, and coordination in Virginia (Terwilliger & observations, strandings, and behavioral activity of Musick, 1995). This study addressed three objectives bottlenose dolphins are well documented at the mouth of under the 1995 management plan for marine mammals: the Chesapeake Bay and along the southern cOastline of (1) to describe the spatial distribution of bottlenose Virginia (Blaylock, 1988: Barco, 1995:, however, few dolphins in the mainstem rivers and tributaries: (2) to data are available on occurrences in mainstem rivers and establish observation programs: and (3) to improve public tributaries of the state. Bottlenose dolphins are known to participation and interest. Similar comprehensive surveys occur in Virginia from April through November (Barco, and long term observation programs were conducted in 1995) and ascend into tributary rivers of the Chesapeake Georgia and Maryland (Wang et al., 1994). -

Evolutionary History of the Porpoises
bioRxiv preprint doi: https://doi.org/10.1101/851469; this version posted November 22, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Evolutionary history of the porpoises (Phocoenidae) across the 2 speciation continuum: a mitogenome phylogeographic perspective 3 4 Yacine Ben Chehida1, Julie Thumloup1, Cassie Schumacher2, Timothy Harkins2, Alex 5 Aguilar3, Asunción Borrell3, Marisa Ferreira4, Lorenzo Rojas-Bracho5, Kelly M. Roberston6, 6 Barbara L. Taylor6, Gísli A. Víkingsson7, Arthur Weyna8, Jonathan Romiguier8, Phillip A. 7 Morin6, Michael C. Fontaine1,9* 8 9 1 Groningen Institute for Evolutionary Life Sciences (GELIFES), University of Groningen, PO Box 11103 CC, 10 Groningen, The Netherlands 11 2 Swift Biosciences, 674 S. Wagner Rd., Suite 100, Ann Arbor, MI 48103, USA 12 3 IRBIO and Department of Evolutive Biology, Ecology and Environmental Sciences, Faculty of Biology, 13 University of Barcelona, Diagonal 643, 08071 Barcelona, Spain 14 4 MATB-Sociedade Portuguesa de Vida Selvagem, Estação de Campo de Quiaios, Apartado EC Quiaios, 3080- 15 530 Figueira da Foz, Portugal & CPRAM-Ecomare, Estrada do Porto de Pesca Costeira, 3830-565 Gafanha da 16 Nazaré, Portugal 17 5 Instituto Nacional de Ecología, Centro de Investigación Científica y de Educación Superior de Ensenada, 18 Carretera Ensenada-Tijuana 3918, Fraccionamiento Zona Playitas, Ensenada, BC 22860, Mexico 19 6 Southwest Fisheries Science Center, National Marine Fisheries Service, NOAA, 8901 La Jolla Shores Dr., La 20 Jolla, California 92037, USA 21 7 Marine and Freshwater Research Institute, PO Box 1390, 121 Reykjavik, Iceland 22 8 Institut des Sciences de l’Évolution (Université de Montpellier, CNRS UMR 5554), Montpellier, France 23 9 Laboratoire MIVEGEC (Université de Montpellier, UMR CNRS 5290, IRD 229), Centre IRD de Montpellier, 24 Montpellier, France 25 26 *Corresponding author: Michael C. -

The Shark-Toothed Dolphin Squalodon Cetacea
Carnets Geol. 20 (2) E-ISSN 1634-0744 DOI 10.4267/2042/70716 The shark-toothed dolphin Squalodon (Cetacea: Odontoceti) from the remarkable Montagna della Majella marine vertebrate assemblage (Bolognano Formation, central Italy) Alberto COLLARETA 1 Andrea DI CENCIO 2 Renato RICCI 3 Giovanni BIANUCCI 4 Abstract: The extinct family Squalodontidae consists of heterodont, medium-sized odontocetes, featu- ring a long rostrum that houses large, procumbent incisors and heavily ornamented postcanine teeth carrying accessory denticles, hence their vernacular name, "shark-toothed dolphins". These longirostri- ne toothed whales are often seen as bridging the anatomical gap between archaic Oligocene odontoce- tes and their late Miocene to Holocene relatives. Possibly among the major marine predators of their ti- me, the shark-toothed dolphins are important components of several lower Miocene marine-mammal assemblages from the North Atlantic and Mediterranean/Paratethysian realms. In the present work, a partial skull of Squalodontidae is described from the strata of the Bolognano Formation cropping out in the northeastern sector of the Montagna della Majella massif (Abruzzo, central Italy), which has pre- viously yielded a rich lower Miocene marine-vertebrate assemblage, including eleven taxa of elasmo- branchs as well as subordinate teleosts and very fragmentary remains of marine reptiles and mam- mals. The specimen consists of the anterodorsal portion of a rostrum, preserving parts of both pre- maxillae and left maxilla, and the anteriormost seven upper left teeth. This partial skull is here identi- fied as belonging to the genus Squalodon, whose presence in the Montagna della Majella vertebrate assemblage had already been tentatively proposed on the basis of two fragmentary teeth. -

Living Between Rapids: Genetic Structure and Hybridization in Botos (Cetacea: Iniidae: Inia Spp.) of the Madeira River, Brazil
bs_bs_banner Biological Journal of the Linnean Society, 2015, 114, 764–777. With 5 figures Living between rapids: genetic structure and hybridization in botos (Cetacea: Iniidae: Inia spp.) of the Madeira River, Brazil WALESKA GRAVENA1,2*, VERA M. F. DA SILVA2, MARIA N. F. DA SILVA3, IZENI P. FARIAS1 and TOMAS HRBEK1 1Laboratório de Evolução e Genética Animal (LEGAL), Departamento de Genética, Universidade Federal do Amazonas, Av. General Rodrigo Otávio Jordão, 3000, 69077-000, Manaus, Amazonas, Brazil 2Laboratório de Mamíferos Aquáticos, Instituto Nacional de Pesquisas da Amazônia, Av. André Araújo, 2936, 69011-970, Manaus, Amazonas, Brazil 3Coleção de Mamíferos, Instituto Nacional de Pesquisas da Amazônia, Av. André Araújo, 2936, 69011-970, Manaus, Amazonas, Brazil Received 2 September 2014; revised 14 November 2014; accepted for publication 15 November 2014 Until the recent construction of hydroelectric dams, a series of 18 rapids divided the upper and lower Madeira River, and these rapids were thought to separate two species of Amazonian freshwater dolphins (boto): Inia boliviensis (above) and I. geoffrensis (below). Some reports and articles, however, mention the occurrence of botos within the rapids region and that they occasionally cross the rapids, but without mentioning the species concerned. Based on our previous studies, it is likely that I. boliviensis occurs in the region of the rapids. To test this supposition, we sampled 18 individuals from this region, and collected mitochondrial (control region, cytochrome b and cytochrome oxidase I) and nuclear (10 microsatellite loci) DNA data, in order to test if there is connectivity between the dolphins that were found within the rapids region and dolphins collected upstream and downstream of the rapids, and investigate population structuring between these localities. -

The Conservation Action Plan the Ganges River Dolphin
THE CONSERVATION ACTION PLAN FOR THE GANGES RIVER DOLPHIN 2010-2020 National Ganga River Basin Authority Ministry of Environment & Forests Government of India Prepared by R. K. Sinha, S. Behera and B. C. Choudhary 2 MINISTER’S FOREWORD I am pleased to introduce the Conservation Action Plan for the Ganges river dolphin (Platanista gangetica gangetica) in the Ganga river basin. The Gangetic Dolphin is one of the last three surviving river dolphin species and we have declared it India's National Aquatic Animal. Its conservation is crucial to the welfare of the Ganga river ecosystem. Just as the Tiger represents the health of the forest and the Snow Leopard represents the health of the mountainous regions, the presence of the Dolphin in a river system signals its good health and biodiversity. This Plan has several important features that will ensure the existence of healthy populations of the Gangetic dolphin in the Ganga river system. First, this action plan proposes a set of detailed surveys to assess the population of the dolphin and the threats it faces. Second, immediate actions for dolphin conservation, such as the creation of protected areas and the restoration of degraded ecosystems, are detailed. Third, community involvement and the mitigation of human-dolphin conflict are proposed as methods that will ensure the long-term survival of the dolphin in the rivers of India. This Action Plan will aid in their conservation and reduce the threats that the Ganges river dolphin faces today. Finally, I would like to thank Dr. R. K. Sinha , Dr. S. K. Behera and Dr. -

Whale Evolution Lab – Page 1
Whale Evolution Lab – Page 1 Whale Evolution Lab Adapted from http://www.indiana.edu/~ensiweb PRELAB (recommended)… FROM LAND TO WATER: A Whale Evolution Internet Activity http://www.indiana.edu/~ensiweb/lessons/whalekiosk.html Click on “SCREEN” to “Find Out More” A. Anatomy B. Fossil record C. Molecular evidence A. WHALE ANATOMY 1. What does the Latin word “cetus” mean? 2. What three groups of organisms are considered cetaceans? 3. What are the two subgroups of cetaceans? 4. What characterizes the subgroup Odontoceti? 5. What characterizes the subgroup Mysticeti? 6. Follow the instructions given to compare anatomical parts. Click on the labels to compare the whale’s anatomy with that of a fish and a cat. Fill out the chart below with your answers to each structure, by placing an “X” under the organism whose structure is more similar to the whale’s. Structure FISH CAT Structure FISH CAT Ears Forelimb Eyes Jaw Lungs Mammary gland 7. According to the anatomical evidence, which organism is more closely related to a whale…fish or cat? 8. Draw and label the cladogram that you created for the whale, fish, and cat below. 9. What is the relationship between whales and cats? B. FOSSIL RECORD 1. What is a fossil? 2. What are the most likely parts to become fossilized? 3. What are trace fossils? List some examples. 4. What is a coprolite? 5. What fossilized anatomical structure can be useful to anatomists? 6. Compare the fossil teeth of whales to the other organisms on the website. What sort of organism has fossil teeth most similar to whale teeth? 7. -

Cetaceans: Whales and Dolphins
CETACEANS: WHALES AND DOLPHINS By Anna Plattner Objective Students will explore the natural history of whales and dolphins around the world. Content will be focused on how whales and dolphins are adapted to the marine environment, the differences between toothed and baleen whales, and how whales and dolphins communicate and find food. Characteristics of specific species of whales will be presented throughout the guide. What is a cetacean? A cetacean is any marine mammal in the order Cetaceae. These animals live their entire lives in water and include whales, dolphins, and porpoises. There are 81 known species of whales, dolphins, and porpoises. The two suborders of cetaceans are mysticetes (baleen whales) and odontocetes (toothed whales). Cetaceans are mammals, thus they are warm blooded, give live birth, have hair when they are born (most lose their hair soon after), and nurse their young. How are cetaceans adapted to the marine environment? Cetaceans have developed many traits that allow them to thrive in the marine environment. They have streamlined bodies that glide easily through the water and help them conserve energy while they swim. Cetaceans breathe through a blowhole, located on the top of their head. This allows them to float at the surface of the water and easily exhale and inhale. Cetaceans also have a thick layer of fat tissue called blubber that insulates their internals organs and muscles. The limbs of cetaceans have also been modified for swimming. A cetacean has a powerful tailfin called a fluke and forelimbs called flippers that help them steer through the water. Most cetaceans also have a dorsal fin that helps them stabilize while swimming. -

Thomas Jefferson Meg Tooth
The ECPHORA The Newsletter of the Calvert Marine Museum Fossil Club Volume 30 Number 3 September 2015 Thomas Jefferson Meg Tooth Features Thomas Jefferson Meg The catalogue number Review; Walking is: ANSP 959 Whales Inside The tooth came from Ricehope Estate, Snaggletooth Shark Cooper River, Exhibit South Carolina. Tiktaalik Clavatulidae In 1806, it was Juvenile Bald Eagle originally collected or Sculpting Whale Shark owned by Dr. William Moroccan Fossils Reid. Prints in the Sahara Volunteer Outing to Miocene-Pliocene National Geographic coastal plain sediments. Dolphins in the Chesapeake Sloth Tooth Found SharkFest Shark Iconography in Pre-Columbian Panama Hippo Skulls CT- Scanned Squalus sp. Teeth Sperm Whale Teeth On a recent trip to the Academy of Natural Sciences of Drexel University (Philadelphia), Collections Manager Ned Gilmore gave John Nance and me a behind -the-scenes highlights tour. Among the fossils that belonged to Thomas☼ Jefferson (left; American Founding Father, principal author of the Declaration of Independence, and third President of the United States) was this Carcharocles megalodon tooth. Jefferson’s interests and knowledge were encyclopedic; a delight to know that they included paleontology. Hand by J. Nance. Photo by S. Godfrey. Jefferson portrait from: http://www.biography.com/people/thomas-jefferson-9353715 ☼ CALVERT MARINE MUSEUM www.calvertmarinemuseum.com 2 The Ecphora September 2015 Book Review: The Walking 41 million years ago and has worldwide distribution. It was fully aquatic, although it did have residual Whales hind limbs. In later chapters, Professor Thewissen George F. Klein discusses limb development and various genetic factors that make whales, whales. This is a The full title of this book is The Walking complicated topic, but I found these chapters very Whales — From Land to Water in Eight Million clear and readable. -

Cetacea: Phocoenidae) from the Upper Part of the Horokaoshirarika Formation (Lower Pliocene), Numata Town, Hokkaido, Japan, and Its Phylogenetic Position
Palaeontologia Electronica palaeo-electronica.org A new skull of the fossil porpoise Numataphocoena yamashitai (Cetacea: Phocoenidae) from the upper part of the Horokaoshirarika Formation (lower Pliocene), Numata Town, Hokkaido, Japan, and its phylogenetic position Yoshihiro Tanaka and Hiroto Ichishima ABSTRACT An early Pliocene porpoise, Numataphocoena yamashitai from Hokkaido, Japan, is known from the holotype, a fairly well-preserved skeleton with an incomplete skull and a referred earbone. A new skull referred to Numataphocoena yamashitai found from almost the same locality as the holotype is interesting because it expands knowl- edge of skull morphology and improves the diagnosis of this taxon. Numataphocoena yamashitai differs from other phocoenids in having the characteristic feature in the maxilla associated with the posterior dorsal infraorbital foramen, narrower and sharper anterior part of the internal acoustic meatus, and a robust anterior process of the peri- otic. A new cladistic analysis places Numataphocoena yamashitai adjacent to Haboro- phocoena toyoshimai and Haborophocoena minutus, among a clade of early branching phocoenids, all of which are chronologically and geographically close to each other. The new skull is probably a younger individual because it is about 80% the size of that of the holotype and it shows closed but unfused sutures. Our description of this specimen helps to understand the intraspecies variation of the extinct species Numataphocoena yamashitai. Yoshihiro Tanaka. Numata Fossil Museum, 2-7-49, Minami 1, Numata Town, Hokkaido, 078-2225 Japan, [email protected] and Hokkaido University Museum, Kita 10, Nishi 8, Kita-ku, Sapporo, Hokkaido 060-0810 Japan Hiroto Ichishima. Fukui Prefectural Dinosaur Museum, Terao 51-11, Muroko, Katsuyama, Fukui 911-8601, Japan, [email protected] Key words: skull; Phocoenidae; phylogeny; maxillary terrace; ontogeny; intraspecies variation Submission: 22 March 2016 Acceptance: 20 October 2016 Tanaka, Yoshihiro and Ichishima, Hiroto. -

Food Consumption and Body Measurements of Amazon River Dolphins (Inia Geoffrensis)
Aquatic Mammals 1999, 25.3, 173–182 Food consumption and body measurements of Amazon river dolphins (Inia geoffrensis) R. A. Kastelein1, B. Neurohr2, S. H. Nieuwstraten1 and P. R. Wiepkema3 1Harderwijk Marine Mammal Park, Strandboulevard Oost 1, 3841 AB Harderwijk, The Netherlands 2Tiergarten Nu¨rnberg, Am Tiergarten 30, D90480 Nu¨rnberg, Germany 3Emeritus Professor of Ethology, Wageningen Agricultural University, Stationsweg 1, 6861 EA Oosterbeek, The Netherlands Abstract dolphin is the most widespread freshwater dolphin in the world, its distribution is limited compared to This report is on the food consumption of 3 male that of most marine odontocetes and it is therefore Amazon river dolphins which were housed in water very likely to become a threatened species. For the of between 27 and 29 C at Duisburg Zoo, management of Amazon river dolphins in the wild, Germany. The food consumption of 2 animals was information is needed about the population size, recorded for 17 successive complete calendar years, age composition and sex ratio, seasonal distribu- that of the third animal for 3 complete successive tion, diet, energy requirements relative to seasonal years. In male 002 the total annual food intake prey distribution and density, and about competi- increased to 1280 kg at the age of 10 years, after tion with other animals and with fisheries. This which it decreased slightly and stabilised at around information could facilitate prey management 1170 kg/year. Male 001 was adult on arrival at the to allow for a certain number of Amazon river zoo. His annual food intake fluctuated at around dolphins in their distribution area. -

4. Palaeontology
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2015 Palaeontology Klug, Christian ; Scheyer, Torsten M ; Cavin, Lionel Posted at the Zurich Open Repository and Archive, University of Zurich ZORA URL: https://doi.org/10.5167/uzh-113739 Conference or Workshop Item Presentation Originally published at: Klug, Christian; Scheyer, Torsten M; Cavin, Lionel (2015). Palaeontology. In: Swiss Geoscience Meeting, Basel, 20 November 2015 - 21 November 2015. 136 4. Palaeontology Christian Klug, Torsten Scheyer, Lionel Cavin Schweizerische Paläontologische Gesellschaft, Kommission des Schweizerischen Paläontologischen Abhandlungen (KSPA) Symposium 4: Palaeontology TALKS: 4.1 Aguirre-Fernández G., Jost J.: Re-evaluation of the fossil cetaceans from Switzerland 4.2 Costeur L., Mennecart B., Schmutz S., Métais G.: Palaeomeryx (Mammalia, Artiodactyla) and the giraffes, data from the ear region 4.3 Foth C., Hedrick B.P., Ezcurra M.D.: Ontogenetic variation and heterochronic processes in the cranial evolution of early saurischians 4.4 Frey L., Rücklin M., Kindlimann R., Klug C.: Alpha diversity and palaeoecology of a Late Devonian Fossillagerstätte from Morocco and its exceptionally preserved fish fauna 4.5 Joyce W.G., Rabi M.: A Revised Global Biogeography of Turtles 4.6 Klug C., Frey L., Rücklin M.: A Famennian Fossillagerstätte in the eastern Anti-Atlas of Morocco: its fauna and taphonomy 4.7 Leder R.M.: Morphometric analysis of teeth of fossil and recent carcharhinid selachiens -
Global Patterns in Marine Mammal Distributions
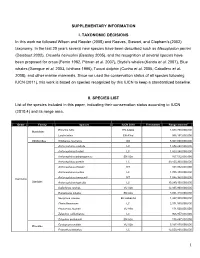
SUPPLEMENTARY INFORMATION I. TAXONOMIC DECISIONS In this work we followed Wilson and Reeder (2005) and Reeves, Stewart, and Clapham’s (2002) taxonomy. In the last 20 years several new species have been described such as Mesoplodon perrini (Dalebout 2002), Orcaella heinsohni (Beasley 2005), and the recognition of several species have been proposed for orcas (Perrin 1982, Pitman et al. 2007), Bryde's whales (Kanda et al. 2007), Blue whales (Garrigue et al. 2003, Ichihara 1996), Tucuxi dolphin (Cunha et al. 2005, Caballero et al. 2008), and other marine mammals. Since we used the conservation status of all species following IUCN (2011), this work is based on species recognized by this IUCN to keep a standardized baseline. II. SPECIES LIST List of the species included in this paper, indicating their conservation status according to IUCN (2010.4) and its range area. Order Family Species IUCN 2010 Freshwater Range area km2 Enhydra lutris EN A2abe 1,084,750,000,000 Mustelidae Lontra felina EN A3cd 996,197,000,000 Odobenidae Odobenus rosmarus DD 5,367,060,000,000 Arctocephalus australis LC 1,674,290,000,000 Arctocephalus forsteri LC 1,823,240,000,000 Arctocephalus galapagoensis EN A2a 167,512,000,000 Arctocephalus gazella LC 39,155,300,000,000 Arctocephalus philippii NT 163,932,000,000 Arctocephalus pusillus LC 1,705,430,000,000 Arctocephalus townsendi NT 1,045,950,000,000 Carnivora Otariidae Arctocephalus tropicalis LC 39,249,100,000,000 Callorhinus ursinus VU A2b 12,935,900,000,000 Eumetopias jubatus EN A2a 3,051,310,000,000 Neophoca cinerea