Complete Plastome Sequences from Bertholletia Excelsa and 23 Related Species Yield Informative Markers for Lecythidaceae
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
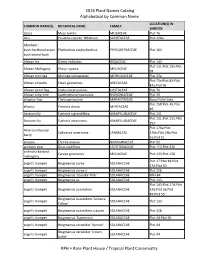
2016 Plant Names Catalog Alphabetical by Common Name
2016 Plant Names Catalog Alphabetical by Common Name LOCATION(S) IN COMMON NAME(S) BOTANICAL NAME FAMILY GARDEN abaca Musa textilis MUSACEAE Plot 76 abiu Pouteria caimito 'Whitman' SAPOTACEAE Plot 128a Abraham- bush:hardhead:scipio- Phyllanthus epiphyllanthus PHYLLANTHACEAE Plot 164 bush:sword-bush African iris Dietes iridioides IRIDACEAE Plot 143 Plot 131:Plot 19a:Plot African Mahogany Khaya nyasica MELIACEAE 58 African moringa Moringa stenopetala MORINGACEAE Plot 32a Plot 71a:Plot 83:Plot African oil palm Elaeis guineensis ARECACEAE 84a:Plot 96 African spiral flag Costus lucanusianus COSTACEAE Plot 76 African tulip-tree Spathodea campanulata BIGNONIACEAE Plot 29 alligator flag Thalia geniculata MARANTACEAE Royal Palm Lake Plot 158:Plot 45:Plot allspice Pimenta dioica MYRTACEAE 46 Amazon lily Eucharis x grandiflora AMARYLLIDACEAE Plot 131 Plot 131:Plot 151:Plot Amazon-lily Eucharis amazonica AMARYLLIDACEAE 152 Plot 176a:Plot American beauty Callicarpa americana LAMIACEAE 176b:Plot 19b:Plot berry 3a:Plot 51 anaqua Ehretia anacua BORAGINACEAE Plot 52 anchovy pear Grias cauliflora LECYTHIDACEAE Plot 112:Plot 32b andiroba:bastard Carapa guianensis MELIACEAE Plot 133:Plot 158 mahogany Plot 17:Plot 18:Plot angel's trumpet Brugmansia aurea SOLANACEAE 27d:Plot 50 angel's trumpet Brugmansia aurea x SOLANACEAE Plot 32b angel's trumpet Brugmansia 'Ecuador Pink' SOLANACEAE RPH-B4 angel's trumpet Brugmansia sp. SOLANACEAE Plot 133 Plot 143:Plot 27d:Plot angel's trumpet Brugmansia suaveolens SOLANACEAE 32b:Plot 3a:Plot 49:Plot 50 Brugmansia suaveolens -

Perennial Edible Fruits of the Tropics: an and Taxonomists Throughout the World Who Have Left Inventory
United States Department of Agriculture Perennial Edible Fruits Agricultural Research Service of the Tropics Agriculture Handbook No. 642 An Inventory t Abstract Acknowledgments Martin, Franklin W., Carl W. Cannpbell, Ruth M. Puberté. We owe first thanks to the botanists, horticulturists 1987 Perennial Edible Fruits of the Tropics: An and taxonomists throughout the world who have left Inventory. U.S. Department of Agriculture, written records of the fruits they encountered. Agriculture Handbook No. 642, 252 p., illus. Second, we thank Richard A. Hamilton, who read and The edible fruits of the Tropics are nnany in number, criticized the major part of the manuscript. His help varied in form, and irregular in distribution. They can be was invaluable. categorized as major or minor. Only about 300 Tropical fruits can be considered great. These are outstanding We also thank the many individuals who read, criti- in one or more of the following: Size, beauty, flavor, and cized, or contributed to various parts of the book. In nutritional value. In contrast are the more than 3,000 alphabetical order, they are Susan Abraham (Indian fruits that can be considered minor, limited severely by fruits), Herbert Barrett (citrus fruits), Jose Calzada one or more defects, such as very small size, poor taste Benza (fruits of Peru), Clarkson (South African fruits), or appeal, limited adaptability, or limited distribution. William 0. Cooper (citrus fruits), Derek Cormack The major fruits are not all well known. Some excellent (arrangements for review in Africa), Milton de Albu- fruits which rival the commercialized greatest are still querque (Brazilian fruits), Enriquito D. -

Recircumscription of the Lecythidaceae
TAXON 47 - NOVEMBER 1998 817 Recircumscription of the Lecythidaceae Cynthia M. Morton'", Ghillean T. Prance', Scott A. Mori4 & Lucy G. Thorburn' Summary Morton. C. M.• Prance, G. T., Mori, S. A. & Thorburn. L. G.: Recircumscriplion of the Le cythidaceae. Taxon 47: 817-827. 1998. -ISSN 004Q-0262. The phylogenetic relationships of the genera of Lecythidaceae and representatives of Scyto petalaceae were assessed using cladistic analysis of both molecular (rbcL and trnL se quences) and morphological data. The results show that the pantropical family Lecythida ceae is paraphyletic. Support was found for the monophyly of three of the four subfamilies: Lecythidoideae, Planchonioideae, and Foetidioideae. The fourth subfamily, Napoleonaeol deae, was found to be paraphyletic, with members of the Scytopetalaceae being nested within it forming a strong clade with Asteranthos. Both families share a number of mor phological features, including several distinct characters such as cortical bundles in the stem. The combined analysis produced three trees of 471 steps and consistency index Cl = 0.71 and retention index Rl = 0.70. Asteranthos !'P.~, members of Scytopetalaceae should be treated as a subfamily of Lecythidaceae, while Napoleonaea and Crateranthus (the latter based solely on morphological features) should remain in the subfamily Napoleo naeoideae.The Lecythldaceaeare recircumscribed, and Asteranthosand members of Scyto peta/aceae are included in Scytopetaloideae. A formal·llWJ!J-pmic synopsis accommodating this new circumscription is presented. Introduction The Lecythidaceae Poit, are 8 pantropical family of trees and shrubs consisting of . 20 genera split into four subfamilies in contemporary classifications (Cronquist, 1981; Prance & Mori, 1979; Takhtajan, 1987; ~ri & Prance, 1990; Thome, 1992). -

Forest Regeneration on the Osa Peninsula, Costa Rica Manette E
University of Connecticut OpenCommons@UConn Master's Theses University of Connecticut Graduate School 12-27-2012 Forest Regeneration on the Osa Peninsula, Costa Rica Manette E. Sandor University of Connecticut, [email protected] Recommended Citation Sandor, Manette E., "Forest Regeneration on the Osa Peninsula, Costa Rica" (2012). Master's Theses. 369. https://opencommons.uconn.edu/gs_theses/369 This work is brought to you for free and open access by the University of Connecticut Graduate School at OpenCommons@UConn. It has been accepted for inclusion in Master's Theses by an authorized administrator of OpenCommons@UConn. For more information, please contact [email protected]. Forest Regeneration on the Osa Peninsula, Costa Rica Manette Eleasa Sandor A.B., Vassar College, 2004 A Thesis Submitted in Partial Fulfillment of the Requirements for the Degree of Master of Science At the University of Connecticut 2012 i APPROVAL PAGE Masters of Science Thesis Forest Regeneration on the Osa Peninsula, Costa Rica Presented by Manette Eleasa Sandor, A.B. Major Advisor________________________________________________________________ Robin L. Chazdon Associate Advisor_____________________________________________________________ Robert K. Colwell Associate Advisor_____________________________________________________________ Michael R. Willig University of Connecticut 2012 ii Acknowledgements Funding for this project was provided through the Connecticut State Museum of Natural History Student Research Award and the Blue Moon Fund. Both Osa Conservation and Lapa Ríos Ecolodge and Wildlife Resort kindly provided the land on which various aspects of the project took place. Three herbaria helpfully provided access to their specimens: Instituto Nacional de Biodiversidad (INBio) in Costa Rica, George Safford Torrey Herbarium at the University of Connecticut, and the Harvard University Herbaria. -

A Phylogeny of Cariniana (Lecythidaceae) Based on Morphological and Anatomical Data
A phylogeny of Cariniana (Lecythidaceae) based on morphological and anatomical data 2 3 YA-YI HUANG!, SCOTT A. MORI , AND GHILLEAN T. PRANCE 1 The New York Botanical Gardenffhe City University of New York, Bronx, New York, NY 10458-5126, USA; e-mail: [email protected] zInstitute of Systematic Botany, The New York Botanical Garden, Bronx, New York, NY 10458 5126, USA; e-mail: [email protected] 3 The Old Vicarage, Silver Street, Lyme Regis, Dorset, UK; e-mail: [email protected] Abstract. Cariniana as previously circumsclibed is a genus of 16 species restricted to neotropical forest habitats on well-drained sites. A phylogenetic analysis of the genus based on 33 morphological and anatomical characters was undertaken. The results show that Cariniana consists of two clades: the AllantomalCariniana decandra clade includes Allantoma lineata and seven species of actinomorphic-flowered Cariniana and is characterized by 5-merous flowers, camose petals, incurved petal apex, scarcely lobed calyces, eucamptodromous secondary veins, dichotomizing venation, and poorly developed areolation; the C. legalis clade is made up of nine species and is characterized by an obliquely zygomorphic androecium, reticulate tertialy venation, and anomocytic stomata. The actinomorphic-flowered Cariniana are more closely re lated to the monotypic Allantoma lineata than they are to the species of the C. legalis clade. In order to reflect these relationships, Cariniana is divided into two genera: species in the C. legalis clade, which includes the generic type C. legalis, remain as Cariniana while species of Cariniana in the AllantomalCariniana decandra clade are transferred to Allantoma. The following new combinations are proposed: AIIantoma decandra, A. -

Table S1. Photographic Records from David Fairchild's 1933
Table S1. Photographic records from David Fairchild’s 1933 expedition to Jamaica. Images are accessioned at the Archives and Library of Fairchild Tropical Botanic Garden (ALFTBG). --------------------------------------------------------------------------------------------------------------- Photograph accession number at ALFTBG / date Photograph description --------------------------------------------------------------------------------------------------------------- 1852 / 3-8-1933 Unknown place. Possibly Castleton Botanical Garden 2230 / 3-8-1933 Tom Barbour besides an akee tree (Blighia sapida K. D. Koenig, Sapindaceae) overlooking stream beside Bath Fountain House 7408 / 3-10-33 Akees in Kingston market 7409 / 3-8-33 Yams (Dioscorea trifida L. f., Dioscoreaceae) in Kingston market 7410 / 3-10-33 Thomas Barbour posing with Geocapromys brownii J. Fisher (Rodentia, Capromyidae). Geocapromys brownii specimen from the John Crow mountains of Jamaica presented by Frank Cundall to Thomas Barbour at Kingston Jamaica. One of these bit James Greenway and he visited a medical doctor to receive a dose of anti- tetanus. See also photograph 7411. Photographs 7410 and 7411 are very similar 7411 / 3-10-33 See photograph 7410. Both photographs are very similar 7418 / 3-10-33 Akees in Kingston market --------------------------------------------------------------------------------------------------------------- Table S1. Continued. --------------------------------------------------------------------------------------------------------------- Photograph -

Observations on the Phytogeography of the Lecythidaceae Clade (Brazil Nut Family)
Mori, S.A., E.A. Kiernan, N.P. Smith, L.M. Kelley, Y-Y. Huang, G.T. Prance & B. Thiers. 2016. Observations on the phytogeography of the Lecythidaceae clade (Brazil nut family). Phytoneuron 2017-30: 1–85. Published 28 April 2017. ISSN 2153 733X OBSERVATIONS ON THE PHYTOGEOGRAPHY OF THE LECYTHIDACEAE CLADE (BRAZIL NUT FAMILY) SCOTT A. MORI Institute of Systematic Botany The New York Botanical Garden Bronx, New York 10458-5126 [email protected] ELIZABETH A. KIERNAN GIS Laboratory The New York Botanical Garden Bronx, New York 10458-5126 NATHAN P. SMITH Research Associate Institute of Systematic Botany The New York Botanical Garden Bronx, New York 10458-5126 LAWRENCE M. KELLY Pfizer Laboratory The New York Botanical Garden Bronx, New York 10458-5126 YA-YI HUANG Biodiversity Research Center Academia Sinica Taipei 11529, Taiwan GHILLEAN T. PRANCE Royal Botanic Gardens Kew, Richmond, Surrey, United Kingdom TW9 3AB BARBARA THIERS Vice President for Science The New York Botanical Garden Bronx, New York 10458-5126 ABSTRACT The Lecythidaceae clade of the order Ericales is distributed in Africa (including Madagascar), Asia in the broadest sense, and South and Central America. Distribution maps are included for the Lecythidaceae clade as follows: family maps for Napoleonaeaceae and Scytopetalaceae; subfamily maps for the Barringtonioideae, Foetidioideae, and Lecythidoideae, and maps for the subclades of Lecythidaceae subfam. Lecythidoideae. The following topics are discussed: (1) the difficulties using herbarium specimens for studies of phytogeography; -

Diversification History of Neotropical Lecythidaceae, an Ecologically
Chapter 29 Diversification History of Neotropical Lecythidaceae, an Ecologically Dominant Tree Family of Amazon Rain Forest Oscar M. Vargas and Christopher W. Dick Abstract The Neotropical subfamily of Lecythidaceae (Lecythidoideae) is a clade of 10 genera with an estimated number of 232 species. Lecythidaceae is the third most abundant family of trees in Amazon forests, and its most diverse genus, Eschweilera (ca. 100 species) is the most abundant genus of Amazon trees. In this chapter we explore the diversification history of the Lecythidoideae through space and time in the Neotropics. We inferred a time-calibrated phylogeny of 118 species, which we used to reconstruct the biogeographic origins of Lecythidoideae and its main clades. To test for significant changes of speciation rates in the subfamily, we performed a diversification analysis. Our analysis dated the crown clade of Lecythidoideae at 46 Ma (95% CI ¼ 36.5–55.9 Ma) and the stem age at 62.7 Ma (95% CI ¼ 56.7–68.9 Ma), suggesting dispersal from the paleotropics long after the Gondwana breakup. Most major crown clades in the Lecythidoideae (Grias, Gustavia, Eschweilera, Couroupita, Couratari, and all Lecythis and Eschweilera subclades) differentiated during the Miocene (ca. 5.3–23 Ma). The Guayana floristic region (Guiana Shield + north-central Amazon) is the inferred ancestral range for 8 out of the 18 Lecythidoideae clades (129 species, ~55%), highlighting the region’s evolutionary importance, especially for the species-rich Bertholletia clade, which includes the genera Eschweilera, Lecythis, Corythophora and Bertholletia. Our results indicate that the Bertholletia clade colonized the Trans-Andean region at least three times in the last 10 Ma. -

THE UTILITY of PLACENTATION in the CIRCUMSCRIPTION of GENERA of NEW WORLD LECYTHIDACEAE (BRAZIL NUT FAMILY) ABSTRACT the Utilit
Mori, S.A., C. Carollo Matos, Y.-Y. Huang, N.P. Smith, and C. de Moraes Potascheff. 2015. The utility of placentation in the circumscription of genera of new world Lecythidaceae (Brazil nut family). Phytoneuron 2015-13: 1–46. Published 3 March 2015. ISSN 2153 733X THE UTILITY OF PLACENTATION IN THE CIRCUMSCRIPTION OF GENERA OF NEW WORLD LECYTHIDACEAE (BRAZIL NUT FAMILY) SCOTT A. MORI Institute of Systematic Botany The New York Botanical Garden Bronx, New York 10458-5126 [email protected] 1 CAROL CAROLLO MATOS Institute of Systematic Botany The New York Botanical Garden Bronx, New York 10458-5126 [email protected] YA-YI HUANG Biodiversity Research Center Academia Sinica Taipei 11529, Taiwan [email protected] NATHAN P. SMITH Institute of Systematic Botany The New York Botanical Garden Bronx, New York 10458-5126 [email protected] C. DE MORAES POTASCHEFF Instituto de Biociência Universidade Estadual Paulista Rio Claro, São Paulo, BraSil Caixa Postal 199, CEP 13506-900 [email protected] ABSTRACT The utility of placentation in the circumscription of genera of New World Lecythidaceae (Brazil nut family). The placentation of the 16 known clades of New World Lecythidaceae and several species of Old World Barringtonia are illustrated and described. The authors conclude that the family possesses axile placentation but an SEM study of flower development may modify that conclusion. They also restrict the term placenta to all tissue derived from the same carpel to which the ovules are attached. The more basal genera are often defined by placental features as well as by morphological apomorphies and sometimes by molecular data. -
Sea Bean Float. Float, Bean, Float. in an Attempt to Expand Their Territory, Some Well-Armored Tropical Seeds and Fruits Hitch Rides on Ocean Currents
SCIENCE Sea bean float. Float, bean, float. In an attempt to expand their territory, some well-armored tropical seeds and fruits hitch rides on ocean currents. These sea beans are often found on our beaches, with more than 150 different species seen from Cape Canaveral to the Keys. A seed pod’s journey across the Atlantic After dropping from their parent trees, many sea beans wash The Gulf Stream, which continues past North America, into inland rivers that flow to the ocean, where they ar e transported takes some seeds to the far-off beaches of northern Europe. by ocean currents to distant beaches. Sea beans have pockets of air that keep them afloat Those that reach shore must overcome compe- for long periods of time, and their seed coat is often tition and foragers, as well as differences in moisture, har d and impermeable to saltwater. Even so, many of temperature and soil conditions, to sprout and survive. these drifters become waterlogged and sink before they reach land. The Gulf Stream current, flowing north from The South Atlantic equatorial current, feeding into the South America and the Gulf of Mexico, deposits Gulf Stream, carries seeds and fruits from Africa, including many sea beans on eastern Florida beaches. the sea heart, true sea-bean, sea purse and gray nickernut. All sea beans shown at actual sizes True sea-bean or hamburger bean Sea-coconut Mucuna spp. Manicaria saccifera Mary’s bean Produced on vines, these Also known as a sleeve palm, this Merremia discoidesperma This seed carries the dis- seeds break out of pods species is native to the American tinctive mark of a cross on that have tiny stinging tropics. -

Lecythidaceae 1745 Mammals (E.G., Squirrels and Monkeys) and fish May Lecythidaceae Also Be Important Dispersers
TROPICAL FORESTS / Lecythidaceae 1745 mammals (e.g., squirrels and monkeys) and fish may Lecythidaceae also be important dispersers. S A Mori, New York Botanical Garden, New York, USA & 2004, Elsevier Ltd. All Rights Reserved. Economic Uses Historically, the Lauraceae is a very important economic family, especially the genus Cinnamomum, Introduction as a source of spices, e.g., cinnamon (C. verum), cassia bark (C. cassia) and laurel (Laurus nobilis), The Lecythidaceae is a pantropical family of trees perfume oils, e.g., rosewood oil (Aniba roseodora) found in the tropics of Central and South America, and sassafras oil (Ocotea odorifera), and pharma- southeast Asia, and Africa, including Madagascar ceuticals, e.g., camphor (C. camphora). However, the (Table 1). The family is divided into five subfamilies: most internationally important product produced (1) the Planchonioideae, with six genera, the best today is the avocado fruit (Persea americana). known of which is Barringtonia, and 59 species in Lauraceae wood is widely used locally, although a tropical Asia, Malaysia, northern Australia, the few are internationally important as high-quality Pacific Islands, and Madagascar; (2) the Foetidioi- timbers for furniture making or for resistance to salt deae, with a single genus, Foetidia, and 17 species in water, e.g., greenheart (Chlorocardium rodiaei), Madagascar, Mauritius, and East Africa; (3) the Borneo ironwood (Eusideroxylon zwageri), and Napoleonaeoideae, with two genera, Crateranthus Queensland walnut (Endiandra palmerstonii). Some and Naopoleonaea, and 11 species in West Africa; (4) species (e.g., Ocotea bullata), once internationally the Scytopetaloideae, with a single species, Aster- important, are now protected because of past over- anthos brasiliensis, in the Negro and Orinoco river exploitation. -

Allergy to Tree Nuts and Edible Seeds
CHAPTER 16 Allergy to Tree Nuts and Edible Seeds ree nuts and edible seeds represent a group of foods that tend to be highly aller- Tgenic and may trigger an anaphylactic reaction in particularly sensitive individu- als. Furthermore, tree nut allergy is especially problematic, because the allergy tends to be persistent and in the majority of cases lasts for a person’s lifetime. Tree Nut Allergy There is not much information on the prevalence of tree nut allergy in populations, and reports vary in the methods of data collection as well as the ages of the study populations. The few reports available indicate that in the United States, an allergy to tree nuts affects about 0.5% of the population.1 In Canada, the number is 1.22%;2 in the United Kingdom, the number varies from 1.4%3 to 2.2%;4 France reports an incidence of 0.7%;5 Germany 8.5%;6 Sweden 4.1%;7 and Israel 0.03%.8 Previously, it was believed that allergies to tree nuts—including cashews, al- monds, Brazil nuts, walnuts, hazelnuts, macadamia nuts, pecans, pistachios, and pine nuts—lasted a lifetime. However, recent research indicates that at least 9% of young children outgrow their nut allergy.9 Even children who have experienced a severe nut allergy can outgrow it. However, the results of the study indicated that children who are allergic to multiple types of tree nuts are less likely to outgrow their allergy than children who are allergic to only one or at most two types of nuts.