Differential Retention of Gene Functions in a Secondary Metabolite Cluster Hannah T
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Diversity and Toxigenicity of Fungi That Cause Pineapple Fruitlet Core Rot
toxins Article Diversity and Toxigenicity of Fungi that Cause Pineapple Fruitlet Core Rot Bastien Barral 1,2,* , Marc Chillet 1,2, Anna Doizy 3 , Maeva Grassi 1, Laetitia Ragot 1, Mathieu Léchaudel 1,4, Noel Durand 1,5, Lindy Joy Rose 6 , Altus Viljoen 6 and Sabine Schorr-Galindo 1 1 Qualisud, Université de Montpellier, CIRAD, Montpellier SupAgro, Univ d’Avignon, Univ de La Reunion, F-34398 Montpellier, France; [email protected] (M.C.); [email protected] (M.G.); [email protected] (L.R.); [email protected] (M.L.); [email protected] (N.D.); [email protected] (S.S.-G.) 2 CIRAD, UMR Qualisud, F-97410 Saint-Pierre, Reunion, France 3 CIRAD, UMR PVBMT, F-97410 Saint-Pierre, Reunion, France; [email protected] 4 CIRAD, UMR Qualisud, F-97130 Capesterre-Belle-Eau, Guadeloupe, France 5 CIRAD, UMR Qualisud, F-34398 Montpellier, France 6 Department of Plant Pathology, Stellenbosch University, Private Bag X1, Matieland 7600, South Africa; [email protected] (L.J.R.); [email protected] (A.V.) * Correspondence: [email protected]; Tel.: +262-2-62-49-27-88 Received: 14 April 2020; Accepted: 14 May 2020; Published: 21 May 2020 Abstract: The identity of the fungi responsible for fruitlet core rot (FCR) disease in pineapple has been the subject of investigation for some time. This study describes the diversity and toxigenic potential of fungal species causing FCR in La Reunion, an island in the Indian Ocean. One-hundred-and-fifty fungal isolates were obtained from infected and healthy fruitlets on Reunion Island and exclusively correspond to two genera of fungi: Fusarium and Talaromyces. -

Alternaria Brassicicola)
Int.J.Curr.Microbiol.App.Sci (2020) 9(8): 2553-2559 International Journal of Current Microbiology and Applied Sciences ISSN: 2319-7706 Volume 9 Number 8 (2020) Journal homepage: http://www.ijcmas.com Original Research Article https://doi.org/10.20546/ijcmas.2020.908.292 In-vivo Management of Alternaria Leaf Spot of Cabbage (Alternaria brassicicola) Dwarkadas T. Bhere*, K. M. Solanke, Amrita Subhadarshini, Shashi Tiwari and Mohan K. Narode Department of Plant Pathology, Sam Higginbottom University of Agriculture, Technology and Sciences, Prayagraj, (U. P), India *Corresponding author ABSTRACT K e yw or ds An experiment was conducted for in-vivo management of Alternaria leaf spot of Cabbage. Alternaria leaf spot, The experiment was analyzed by using RBD (randomized block design) with three 2 Cabbage, replications in a plot size 2x2m . Eight treatments were taken i.e. Neem oil, Eucalyptus oil, Eucalyptus oil and Clove oil, Trichoderma viride, Neem oil + Trichoderma viride, Eucalyptus oil + Clove oil, Trichoderma viride, Clove oil + Trichoderma viride along with the control. Observations Trichoderma viride, were recorded at disease intensity 30, 45 and 60 (days after Transplanting), plant growth Neem oil parameters such a yield (q/ha). Experiment revealed that Neem oil significantly reduced the Alternaria leaf spot of Cabbage, where among the use Neem oil seedling treatment @ Article Info 5% increased the yield. The maximum cost benefit ratio was recorded by Neem oil (1:3.26) Thus according to experimental finding and results discussed in the earlier Accepted: chapter, it is concluded that Neem oil reduced the Alternaria leaf spot of Cabbage, where 22 July 2020 among the Neem oil seedling application found maximum yield was significantly superior Available Online: 10 August 2020 as compare to other treatments. -

Infection Cycle of Alternaria Brassicicola on Brassica Oleracea Leaves Under Growth Room Conditions
Plant Pathology (2018) 67, 1088–1096 Doi: 10.1111/ppa.12828 Infection cycle of Alternaria brassicicola on Brassica oleracea leaves under growth room conditions V. K. Macioszeka, C. B. Lawrenceb and A. K. Kononowicza* aDepartment of Genetics, Plant Molecular Biology and Biotechnology, Faculty of Biology and Environmental Protection, University of Lodz, 90-237 Lodz, Poland; and bDepartment of Biological Sciences, Virginia Tech, Blacksburg, VA 24061, USA Development of the necrotrophic fungus Alternaria brassicicola was evaluated during infection of three cabbage vari- eties: Brassica oleracea var. capitata f. alba ‘Stone Head’ (white cabbage), B. oleracea var. capitata f. rubra ‘Langedi- jker Dauer’ (red cabbage) and B. oleracea var. capitata f. sabauda ‘Langedijker Dauerwirsing’ (Savoy cabbage). Following inoculation of cabbage leaves, conidial germination, germ tube growth, and appressorium formation were analysed during the first 24 h of infection. Differences in the dynamics of fungal development on leaves were observed, e.g. approximately 40% of conidia germinated on Savoy cabbage leaves at 4 h post-inoculation (hpi) while only 20% germinated on red and white cabbage leaves. Leaf penetration on the three cabbage varieties mainly occurred through appressoria, rarely through stomata. Formation of infection cushions was found exclusively on red cabbage. Appresso- ria were first observed on red cabbage leaves at 6 hpi, and on white and Savoy cabbage leaves at 8 hpi. Conidiogenesis occurred directly from mature conidia at an early stage of fungal development (10 hpi), but later (48 hpi) it occurred through conidiophores. Disease progress and changes in the morphology of leaf surfaces were also observed. At the final 120 hpi measurement point, necroses on all investigated varieties were approximately the same size. -

Nature and Effect of Alternaria Spp. Complex from Wheat Grain on Germination and Disease Transmission
Pak. J. Bot., 45(5): 1817-1824, 2013. NATURE AND EFFECT OF ALTERNARIA SPP. COMPLEX FROM WHEAT GRAIN ON GERMINATION AND DISEASE TRANSMISSION ANALÍA E. PERELLÓ1,2* AND SILVINA LARRÁN1 1CIDEFI (Centro de Investigaciones de Fitopatología) y Cátedra de Fitopatología 2CONICET-Facultad de Ciencias Agrarias y Forestales de la Universidad Nacional de La Plata, Calle 60 y 119 (1900) La Plata, Buenos Aires, Argentina. *Corresponding author’s e-mail: anaperello2@ yahoo.com.ar Abstract Diseases caused by Alternaria sp. are among the most common diseases of crops throughout the world. Alternaria sp. is a common component of the flora of wheat seed. Although isolation of Alternaria sp. from wheat (Triticum aestivum) seed has been reported in Argentina, development of the Alternaria blight in plants from infected seeds has not been demonstrated experimentally. Seed transmission of strains belonging to Alternaria tenuissima, A. alternata, A. infectoria, A. triticina, A. chlamydospora and related genera like Embellisia and Ulocladium sp. on wheat were investigated in the Argentinean growing area, on wheat cultivars Klein Escorpión and Buck Poncho. A. tenuissima was the dominant fungus in black pointed kernels. Transmission of all 42 seed-borne members of Alternaria complex from seeds to seedlings artificially inoculated was detected by trays seedling symptoms test. Among the fungi tested most isolates of Alternaria, Embellisia sp. and Ulocladium sp. produced distinct seed rot and seedling infection symptoms. This confirmed the seed-borne nature of these fungi. In each wheat cultivar tested inoculated seeds appreciably reduced their germination. The emerging coleoptile is externally infected by hyphal growth from the infected pericarp. -
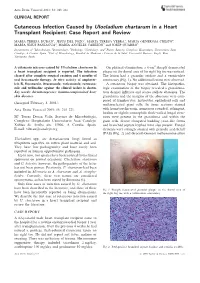
Cutaneous Infection Caused by Ulocladium Chartarum in a Heart Transplant Recipient: Case Report and Review
Acta Derm Venereol 2003; 83: 218–221 CLINICAL REPORT Cutaneous Infection Caused by Ulocladium chartarum in a Heart Transplant Recipient: Case Report and Review MARI´A TERESA DURA´ N1, JESU´ S DEL POZO2, MARI´A TERESA YEBRA3, MARI´A GENEROSA CRESPO4, MARI´A JESU´ S PANIAGUA4, MARI´A ANGELES CABEZO´ N5 and JOSEP GUARRO6 Departments of 1Microbiology, 2Dermatology, 3Pathology, 4Cardiology, and 5Plastic Surgery, Complexo Hospitalario Universitario Juan Canalejo, A Corun˜a, Spain, 6Unit of Microbiology, Facultat de Medicina i Cie`ncies de la Salut, Universitat Rovira i Virgili, Reus, Tarragona, Spain A cutaneous mycoses caused by Ulocladium chartarum in On physical examination, a 6-cm2 sharply demarcated a heart transplant recipient is reported. The infection plaque on the dorsal area of his right big toe was noticed. cleared after complete surgical excision and 6 months of The lesion had a granular surface and a vermiculate oral itraconazole therapy. In vitro activity of amphoter- consistency (Fig. 1). No additional lesions were observed. icin B, fluconazole, itraconazole, voriconazole, ravucona- A cutaneous biopsy was obtained. The histopatho- zole and terbinafine against the clinical isolate is shown. logic examination of the biopsy revealed a granuloma- Key words: dermatomycoses; immunocompromised host; tous dermal infiltrate and scarce stellate abscesses. The skin diseases. granuloma and the margins of the abscesses were com- posed of lymphocytes, histiocytes, epithelioid cells and (Accepted February 3, 2003.) multinucleated giant cells. In tissue sections stained Acta Derm Venereol 2003; 83: 218–221. with hematoxylin-eosin, numerous rounded, refringent, hyaline or slightly eosinophilic thick-walled fungal struc- Ma Teresa Dura´n Valle, Servicio de Microbiologı´a, tures were present in the granuloma and within the Complexo Hospitalario Universitario Juan Canalejo, giant cells. -

The Emergence of Cereal Fungal Diseases and the Incidence of Leaf Spot Diseases in Finland
AGRICULTURAL AND FOOD SCIENCE AGRICULTURAL AND FOOD SCIENCE Vol. 20 (2011): 62–73. Vol. 20(2011): 62–73. The emergence of cereal fungal diseases and the incidence of leaf spot diseases in Finland Marja Jalli, Pauliina Laitinen and Satu Latvala MTT Agrifood Research Finland, Plant Production Research, FI-31600 Jokioinen, Finland, email: [email protected] Fungal plant pathogens causing cereal diseases in Finland have been studied by a literature survey, and a field survey of cereal leaf spot diseases conducted in 2009. Fifty-seven cereal fungal diseases have been identified in Finland. The first available references on different cereal fungal pathogens were published in 1868 and the most recent reports are on the emergence of Ramularia collo-cygni and Fusarium langsethiae in 2001. The incidence of cereal leaf spot diseases has increased during the last 40 years. Based on the field survey done in 2009 in Finland, Pyrenophora teres was present in 86%, Cochliobolus sativus in 90% and Rhynchosporium secalis in 52% of the investigated barley fields.Mycosphaerella graminicola was identi- fied for the first time in Finnish spring wheat fields, being present in 6% of the studied fields.Stagonospora nodorum was present in 98% and Pyrenophora tritici-repentis in 94% of spring wheat fields. Oat fields had the fewest fungal diseases. Pyrenophora chaetomioides was present in 63% and Cochliobolus sativus in 25% of the oat fields studied. Key-words: Plant disease, leaf spot disease, emergence, cereal, barley, wheat, oat Introduction nbrock and McDonald 2009). Changes in cropping systems and in climate are likely to maintain the plant-pathogen interactions (Gregory et al. -

Identification and Characterization of Alternaria Species Causing Leaf Spot
Eur J Plant Pathol (2017) 149:401–413 DOI 10.1007/s10658-017-1190-0 Identification and characterization of Alternaria species causing leaf spot on cabbage, cauliflower, wild and cultivated rocket by using molecular and morphological features and mycotoxin production Ilenia Siciliano & Giovanna Gilardi & Giuseppe Ortu & Ulrich Gisi & Maria Lodovica Gullino & Angelo Garibaldi Accepted: 22 February 2017 /Published online: 11 March 2017 # Koninklijke Nederlandse Planteziektenkundige Vereniging 2017 Abstract Alternaria species are common pathogens of Keywords Crucifers . Toxins . Leaf spot . Tenuazonic fruit and vegetables able to produce secondary metabo- acid lites potentially affecting human health. Twenty-nine isolates obtained from cabbage, cauliflower, wild and cultivated rocket were characterized and identified Introduction based on sporulation pattern and virulence; the phylo- β genetic analysis was based on the -tubulin gene. Most Alternaria species are saprophytes and ubiquitous Isolates were identified as A. alternata, A. tenuissima, in the environment, however some are plant pathogenic, A. arborescens, A. brassicicola and A. japonica. inducing diseases on a large variety of economically Pathogenicity was evaluated on plants under greenhouse important crops like cereals, oil-crops, vegetables and conditions. Two isolates showed low level of virulence fruits (Pitt and Hocking, 1997). Most Alternaria spp. on cultivated rocket while the other isolates showed produce chains of conidia with transverse and longitu- medium or high level of virulence. Isolates were also dinal septa with a tapering apical cell. Conidial size, characterized for their mycotoxin production on a mod- presence and size of a beak, the pattern of catenation ified Czapek-Dox medium. Production of the five and longitudinal and transverse septation are key taxo- Alternaria toxins, tenuazonic acid, alternariol, nomic features for this genus (Joly 1964; Ellis 1971 and alternariol monomethyl ether, altenuene and tentoxin 1976, Simmons 1992). -

Fusarium Langsethiae on Kernels of Winter Wheat in Poland – Occurrence and Mycotoxigenic Abilities
3rd Int. FHB Symposium Szeged, Hungary, 2008 FUSARIUM LANGSETHIAE ON KERNELS OF WINTER WHEAT IN POLAND – OCCURRENCE AND MYCOTOXIGENIC ABILITIES Aleksander LUKANOWSKI – Czeslaw SADOWSKI Department of Phytopathology, University of Technology and Life Sciences, Ks. A. Kordeckiego 20, 85-225 Bydgoszcz, Poland, e-mail: [email protected] Abstract: The object of the study was mycological analysis of settlement of winter wheat grain samples collected in 2005/2006 and 2006/2007 cropping seasons with the special respect to a new species – Fusarium langsethiae. Its presence was noted in 38 samples in relatively low intensity (from 0.5 up to 3.5%). PCR assay confirmed identity of 38 isolates as F. langsethiae. Mycelium of tested strains of F. langsethiae was whitish with powdery appearance. Microconidia were formed napiform or globose, nonseptate, formed in heads. Macroconidia, sclerotia, and chlamydospores were not present after 3 weeks of incubation. Growth rates ranged from 5.4 to 10.3 mm/day. No isolate had potential ability to type B trichothecenes. Detailed chemical analyses of two strains showed them as producers of T-2 and HT-2 toxins at high concentrations. Key words: Fusarium langsethiae, PCR, wheat, mycotoxins Introduction Fungi from genus Fusarium, one of the most dangerous pathogens of cereals, cause serious loses of yield and significantly decrease its quality and wheat shows relatively high susceptibility to infection by these fungi. During research on mycotoxins and the occurrence of Fusarium spp. in cereal grain in Norway (Kosiak et al. 1997; Langseth and Rundberget 1999), it was found a Fusarium species of uncertain identity morphologically resembling F. -

Delimitation of Neonectria and Cylindrocarpon (Nectriaceae, Hypocreales, Ascomycota) and Related Genera with Cylindrocarpon-Like Anamorphs
available online at www.studiesinmycology.org StudieS in Mycology 68: 57–78. 2011. doi:10.3114/sim.2011.68.03 Delimitation of Neonectria and Cylindrocarpon (Nectriaceae, Hypocreales, Ascomycota) and related genera with Cylindrocarpon-like anamorphs P. Chaverri1*, C. Salgado1, Y. Hirooka1, 2, A.Y. Rossman2 and G.J. Samuels2 1University of Maryland, Department of Plant Sciences and Landscape Architecture, 2112 Plant Sciences Building, College Park, Maryland 20742, USA; 2United States Department of Agriculture, Agriculture Research Service, Systematic Mycology and Microbiology Laboratory, Rm. 240, B-010A, 10300 Beltsville Avenue, Beltsville, Maryland 20705, USA *Correspondence: Priscila Chaverri, [email protected] Abstract: Neonectria is a cosmopolitan genus and it is, in part, defined by its link to the anamorph genusCylindrocarpon . Neonectria has been divided into informal groups on the basis of combined morphology of anamorph and teleomorph. Previously, Cylindrocarpon was divided into four groups defined by presence or absence of microconidia and chlamydospores. Molecular phylogenetic analyses have indicated that Neonectria sensu stricto and Cylindrocarpon sensu stricto are phylogenetically congeneric. In addition, morphological and molecular data accumulated over several years have indicated that Neonectria sensu lato and Cylindrocarpon sensu lato do not form a monophyletic group and that the respective informal groups may represent distinct genera. In the present work, a multilocus analysis (act, ITS, LSU, rpb1, tef1, tub) was applied to representatives of the informal groups to determine their level of phylogenetic support as a first step towards taxonomic revision of Neonectria sensu lato. Results show five distinct highly supported clades that correspond to some extent with the informal Neonectria and Cylindrocarpon groups that are here recognised as genera: (1) N. -

Fusarium-Produced Mycotoxins in Plant-Pathogen Interactions
toxins Review Fusarium-Produced Mycotoxins in Plant-Pathogen Interactions Lakshmipriya Perincherry , Justyna Lalak-Ka ´nczugowska and Łukasz St˛epie´n* Plant-Pathogen Interaction Team, Department of Pathogen Genetics and Plant Resistance, Institute of Plant Genetics, Polish Academy of Sciences, Strzeszy´nska34, 60-479 Pozna´n,Poland; [email protected] (L.P.); [email protected] (J.L.-K.) * Correspondence: [email protected] Received: 29 October 2019; Accepted: 12 November 2019; Published: 14 November 2019 Abstract: Pathogens belonging to the Fusarium genus are causal agents of the most significant crop diseases worldwide. Virtually all Fusarium species synthesize toxic secondary metabolites, known as mycotoxins; however, the roles of mycotoxins are not yet fully understood. To understand how a fungal partner alters its lifestyle to assimilate with the plant host remains a challenge. The review presented the mechanisms of mycotoxin biosynthesis in the Fusarium genus under various environmental conditions, such as pH, temperature, moisture content, and nitrogen source. It also concentrated on plant metabolic pathways and cytogenetic changes that are influenced as a consequence of mycotoxin confrontations. Moreover, we looked through special secondary metabolite production and mycotoxins specific for some significant fungal pathogens-plant host models. Plant strategies of avoiding the Fusarium mycotoxins were also discussed. Finally, we outlined the studies on the potential of plant secondary metabolites in defense reaction to Fusarium infection. Keywords: fungal pathogens; Fusarium; pathogenicity; secondary metabolites Key Contribution: The review summarized the knowledge and recent reports on the involvement of Fusarium mycotoxins in plant infection processes, as well as the consequences for plant metabolism and physiological changes related to the pathogenesis. -

Managing Alternaria Brown Spot by L.W
Managing Alternaria brown spot By L.W. Timmer, R. F. Reis, S.N. Mondal and N.A. Peres anker and greening may be problem for production of Fortune tan - the diseases that are on gerine in Spain. More recently, we Fig. 1. Alternaria brown spot lesions everyone’s minds, but we have confirmed the presence of the on a Minneola tangelo leaf. Note the still have to deal with the disease in Peru, where it is severe on characteristic necrosis running up Cmany everyday problems of produc - Minneolas, and in Iran, where it the veins. tion. Alternaria brown spot is the most causes problems mostly for Fortune for a few days on the grove floor, but serious disease of many popular tan - and Page production. then ceases as the leaves decay. Spores gerines and tangerine hybrids such as Tangerine cultivars, including Min - can also be produced on fruit and Minneola and Orlando tangelos, Mur - neolas, Orlandos, Novas, Lees, twigs, but are relatively few compared cotts, Sunburst, Novas and others. Ponkan, Murcotts, Dancy and many to the production on leaves. However, This disease probably requires more others, that are susceptible to ABS lesions on fruit and twigs may be im - intense management than any of the carry a gene for susceptibility to the portant in the overw-inter survival of other fungal diseases. toxin. That gene is dominant and all of the pathogen. Alternaria brown spot (ABS) is the offspring of susceptible parents are In the 2004-05 season, Alternaria caused by the fungus Alternaria alter - also susceptible. Many cultivars devel - pressure was very low and growers nata . -

Phylogenetic Analyses and Toxigenic Profiles of Fusarium Equiseti and Fusarium Acuminatum Isolated from Cereals from Southern Eu
Phylogenetic analyses and toxigenic profiles of Fusarium equiseti and Fusarium acuminatum isolated from cereals from Southern Europe Patricia Marin a, Antonio Morettib, Alberto Ritienic, Miguel Jurado d, Covadonga Vazquez e, M. Teresa Gonzalez-Jaena* ABSTRACT Fusarium equiseti and Fusarium acuminatum are toxigenic species that contaminate cereal crops from diverse climatic regions. They are common in Spanish cereals. The information available on their phylogenetics and toxigenic profiles is, however, insufficient to assist risk evaluation. In this work, phylogenetic analyses were performed using partial sequences of the translation elongation factor gene (EF-\a) of F. equiseti and F. acuminatum strains isolated from barley and wheat from Spain and other countries. The Northern and Southern European F. equiseti strains largely separated into two phyloge- netically distinct clusters. This suggests the existence of two distinct populations within this species, explaining its presence in these regions of markedly different climate. Production of type A and B trichothecenes by the Spanish strains, examined in wheat cultures using a multitoxin analytical method, indicated that F. equiseti could produce deoxynivalenol and nivalenol and other trichothecenes, at concentrations that might represent a significant risk of toxin contamination for Southern European cereals. F. acuminatum showed low intraspecific genetic variability and 58% of the strains could produce deoxynivalenol at low level. Neither species was found to produce T-2 or HT-2 toxins. The present results provide important phylogenetic and toxigenic information essential for the accurate prediction of toxigenic risk. 1. Introduction Fusarium acuminatum, Fusarium subglutinans, Fusarium solani, Fusarium oxysporum, Fusarium semitectum, Fusarium verticillioides Cereals are a dietary staple in most temperate regions.