Phototransduction Cascade Activation Steps
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Reprogramming Metabolism by Targeting Sirtuin 6 Attenuates Retinal Degeneration
The Journal of Clinical Investigation RESEARCH ARTICLE Reprogramming metabolism by targeting sirtuin 6 attenuates retinal degeneration Lijuan Zhang,1,2,3 Jianhai Du,4 Sally Justus,1,2 Chun-Wei Hsu,1,2 Luis Bonet-Ponce,5 Wen-Hsuan Wu,1,2 Yi-Ting Tsai,1,2 Wei-Pu Wu,1,2 Yading Jia,2 Jimmy K. Duong,6 Vinit B. Mahajan,7 Chyuan-Sheng Lin,8 Shuang Wang,6 James B. Hurley,4 and Stephen H. Tsang1,2,9 1The Jonas Children’s Vision Care and Bernard & Shirlee Brown Glaucoma Laboratory, Department of Ophthalmology, Columbia University, New York, New York, USA. 2Edward S. Harkness Eye Institute, New York-Presbyterian Hospital, New York, New York, USA. 3Shanxi Eye Hospital, affiliated with Shanxi Medical University, Xinghualing, Taiyuan, China. 4Department of Biochemistry and Ophthalmology, University of Washington, Seattle, Washington, USA. 5Section on Retinal Ganglion Cell Biology, Laboratory of Retinal Cell and Molecular Biology, NIH, Bethesda, Maryland, USA. 6Department of Biostatistics, Mailman School of Public Health, Columbia University Medical Center, New York, New York, USA. 7Omics Lab, University of Iowa, Iowa City, Iowa, USA. 8Department of Pathology and Cell Biology, Herbert Irving Comprehensive Cancer Center, College of Physicians and Surgeons of Columbia University, New York, New York, USA. 9Departments of Pathology and Cell Biology, Institute of Human Nutrition, College of Physicians and Surgeons, Columbia University, New York, New York, USA. Retinitis pigmentosa (RP) encompasses a diverse group of Mendelian disorders leading to progressive degeneration of rods and then cones. For reasons that remain unclear, diseased RP photoreceptors begin to deteriorate, eventually leading to cell death and, consequently, loss of vision. -
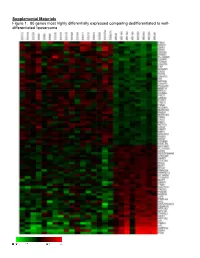
Supplemental Materials Figure 1. 80 Genes Most Highly Differentially Expressed Comparing Dedifferentiated to Well- Differentiated Liposarcoma
Supplemental Materials Figure 1. 80 genes most highly differentially expressed comparing dedifferentiated to well- differentiated liposarcoma Figure 2. Validation of microa CDC2, RACGAP1, FGFR-2, MAD2 and CITED1 200 CDK4 by Liposarcoma Subtype 16 0 12 0 80 Expression40 Level 0 Nl Fat rray data by quantitative RT-P Well Diff 16 0 p16 by Liposarcoma Subtype Dediff 12 0 80 Myxoid Expression40 Level Round Cell 0 12 0 MDM2 by Liposarcoma Subtype 10 0 Pleomorphic Nl Fat 80 60 40 Expression Level 20 RACGAP1 by LiposarcomaWell Diff Subtype 0 70 CR for genes CDK4, MDM2, p16, 60 Dediff 50 Nl Fat 40 30 Myxoid Expression20 Level Well Diff 10 50 0 Round Cell CDC2 by Liposarcoma Subtype Dediff 40 Nl Fat Pleomorphic 30 Myxoid 20 Expression Level Well Diff 10 MAD2L1 by Liposarcoma Subtype Round Cell 60 0 50 Dediff Pleomorphic 40 Nl Fat 30 Myxoid 20 Expression Level Well Diff 10 FGFR-2 by Liposarcoma Subtype 0 Round Cell 200 16 0 Dediff Nl Fat Pleomorphic 12 0 Myxoid 80 Expression Level Well Diff 40 Round Cell 0 Dediff Pleomorphic Nl Fat Myxoid Well Diff CITED1 by Liposarcoma Subtype Round Cell 10 0 80 Dediff Pleomorphic 60 40 Myxoid Expression Level 20 0 Round Cell Nl Fat Pleomorphic Well Diff Dediff Myxoid Round Cell Pleomorphic Table 1. 142 Gene classifier for liposarcoma subtype The genes used for each pair wise subtype comparison are grouped together. The flag column indicates which genes are unique to each subtype comparison. The values show the mean expression levels (actually the mean of the log expression levels was computed and than transformed back to absolute expression level). -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

The Capacity of Long-Term in Vitro Proliferation of Acute Myeloid
The Capacity of Long-Term in Vitro Proliferation of Acute Myeloid Leukemia Cells Supported Only by Exogenous Cytokines Is Associated with a Patient Subset with Adverse Outcome Annette K. Brenner, Elise Aasebø, Maria Hernandez-Valladares, Frode Selheim, Frode Berven, Ida-Sofie Grønningsæter, Sushma Bartaula-Brevik and Øystein Bruserud Supplementary Material S2 of S31 Table S1. Detailed information about the 68 AML patients included in the study. # of blasts Viability Proliferation Cytokine Viable cells Change in ID Gender Age Etiology FAB Cytogenetics Mutations CD34 Colonies (109/L) (%) 48 h (cpm) secretion (106) 5 weeks phenotype 1 M 42 de novo 241 M2 normal Flt3 pos 31.0 3848 low 0.24 7 yes 2 M 82 MF 12.4 M2 t(9;22) wt pos 81.6 74,686 low 1.43 969 yes 3 F 49 CML/relapse 149 M2 complex n.d. pos 26.2 3472 low 0.08 n.d. no 4 M 33 de novo 62.0 M2 normal wt pos 67.5 6206 low 0.08 6.5 no 5 M 71 relapse 91.0 M4 normal NPM1 pos 63.5 21,331 low 0.17 n.d. yes 6 M 83 de novo 109 M1 n.d. wt pos 19.1 8764 low 1.65 693 no 7 F 77 MDS 26.4 M1 normal wt pos 89.4 53,799 high 3.43 2746 no 8 M 46 de novo 26.9 M1 normal NPM1 n.d. n.d. 3472 low 1.56 n.d. no 9 M 68 MF 50.8 M4 normal D835 pos 69.4 1640 low 0.08 n.d. -

Predicting Coupling Probabilities of G-Protein Coupled Receptors Gurdeep Singh1,2,†, Asuka Inoue3,*,†, J
Published online 30 May 2019 Nucleic Acids Research, 2019, Vol. 47, Web Server issue W395–W401 doi: 10.1093/nar/gkz392 PRECOG: PREdicting COupling probabilities of G-protein coupled receptors Gurdeep Singh1,2,†, Asuka Inoue3,*,†, J. Silvio Gutkind4, Robert B. Russell1,2,* and Francesco Raimondi1,2,* 1CellNetworks, Bioquant, Heidelberg University, Im Neuenheimer Feld 267, 69120 Heidelberg, Germany, 2Biochemie Zentrum Heidelberg (BZH), Heidelberg University, Im Neuenheimer Feld 328, 69120 Heidelberg, Germany, 3Graduate School of Pharmaceutical Sciences, Tohoku University, Sendai, Miyagi 980-8578, Japan and 4Department of Pharmacology and Moores Cancer Center, University of California, San Diego, La Jolla, CA 92093, USA Received February 10, 2019; Revised April 13, 2019; Editorial Decision April 24, 2019; Accepted May 01, 2019 ABSTRACT great use in tinkering with signalling pathways in living sys- tems (5). G-protein coupled receptors (GPCRs) control multi- Ligand binding to GPCRs induces conformational ple physiological states by transducing a multitude changes that lead to binding and activation of G-proteins of extracellular stimuli into the cell via coupling to situated on the inner cell membrane. Most of mammalian intra-cellular heterotrimeric G-proteins. Deciphering GPCRs couple with more than one G-protein giving each which G-proteins couple to each of the hundreds receptor a distinct coupling profile (6) and thus specific of GPCRs present in a typical eukaryotic organism downstream cellular responses. Determining these coupling is therefore critical to understand signalling. Here, profiles is critical to understand GPCR biology and phar- we present PRECOG (precog.russelllab.org): a web- macology. Despite decades of research and hundreds of ob- server for predicting GPCR coupling, which allows served interactions, coupling information is still missing for users to: (i) predict coupling probabilities for GPCRs many receptors and sequence determinants of coupling- specificity are still largely unknown. -

Impedes Transport of GRK1 and PDE6 Catalytic Subunits to Photoreceptor Outer Segments
Deletion of PrBP/␦ impedes transport of GRK1 and PDE6 catalytic subunits to photoreceptor outer segments H. Zhang*, S. Li*, T. Doan†, F. Rieke†‡, P. B. Detwiler†, J. M. Frederick*, and W. Baehr*§¶ʈ *John A. Moran Eye Center, University of Utah Health Science Center, Salt Lake City, UT 84132; †Department of Physiology and Biophysics and ‡Howard Hughes Medical Institute, University of Washington, Seattle, WA 98195; and Departments of §Neurobiology and Anatomy and ¶Biology, University of Utah, Salt Lake City, UT 84112 Edited by Jeremy Nathans, Johns Hopkins University School of Medicine, Baltimore, MD, and approved April 11, 2007 (received for review February 23, 2007) The mouse Pde6d gene encodes a ubiquitous prenyl binding chains (5, 7). Posttranslational sorting and targeting of proteins protein, termed PrBP/␦, of largely unknown physiological function. occurs in all cells and is of particular importance in photore- PrBP/␦ was originally identified as a putative rod cGMP phospho- ceptors, which renew their entire outer segments roughly every diesterase (PDE6) subunit in the retina, where it is relatively 10 days (8). Because of compartmentalization of inner and outer abundant. To investigate the consequences of Pde6d deletion in segments and very active metabolism, photoreceptors are re- .retina, we generated a Pde6d؊/؊ mouse by targeted recombina- garded as model cells to study protein trafficking tion. Although manifesting reduced body weight, the Pde6d؊/؊ PrBP/␦, originally thought to be a subunit of PDE6 and termed mouse was viable and fertile and its retina developed normally. PDE␦ (9), was shown recently to be a prenyl binding protein (10, Immunocytochemistry showed that farnesylated rhodopsin kinase 11) and subsequently named PrBP/␦ to reflect this fact (12). -

G-Protein ␥-Complex Is Crucial for Efficient Signal Amplification in Vision
The Journal of Neuroscience, June 1, 2011 • 31(22):8067–8077 • 8067 Cellular/Molecular G-Protein ␥-Complex Is Crucial for Efficient Signal Amplification in Vision Alexander V. Kolesnikov,1 Loryn Rikimaru,2 Anne K. Hennig,1 Peter D. Lukasiewicz,1 Steven J. Fliesler,4,5,6,7 Victor I. Govardovskii,8 Vladimir J. Kefalov,1 and Oleg G. Kisselev2,3 1Department of Ophthalmology and Visual Sciences, Washington University School of Medicine, St. Louis, Missouri 63110, Departments of 2Ophthalmology and 3Biochemistry and Molecular Biology, Saint Louis University School of Medicine, Saint Louis, Missouri 63104, 4Research Service, Veterans Administration Western New York Healthcare System, and Departments of 5Ophthalmology (Ross Eye Institute) and 6Biochemistry, University at Buffalo/The State University of New York (SUNY), and 7SUNY Eye Institute, Buffalo, New York 14215, and 8Sechenov Institute for Evolutionary Physiology and Biochemistry, Russian Academy of Sciences, Saint Petersburg 194223, Russia A fundamental question of cell signaling biology is how faint external signals produce robust physiological responses. One universal mechanism relies on signal amplification via intracellular cascades mediated by heterotrimeric G-proteins. This high amplification system allows retinal rod photoreceptors to detect single photons of light. Although much is now known about the role of the ␣-subunit of the rod-specific G-protein transducin in phototransduction, the physiological function of the auxiliary ␥-complex in this process remains a mystery. Here, we show that elimination of the transducin ␥-subunit drastically reduces signal amplification in intact mouse rods. The consequence is a striking decline in rod visual sensitivity and severe impairment of nocturnal vision. Our findings demonstrate that transducin ␥-complex controls signal amplification of the rod phototransduction cascade and is critical for the ability of rod photoreceptors to function in low light conditions. -

It Takes Two Transducins to Activate the Cgmp-Phosphodiesterase 6 in Retinal Rods Rsob.Royalsocietypublishing.Org Bilal M
It takes two transducins to activate the cGMP-phosphodiesterase 6 in retinal rods rsob.royalsocietypublishing.org Bilal M. Qureshi1,†,‡, Elmar Behrmann1,†,}, Johannes Scho¨neberg2,jj, Justus Loerke1,Jo¨rg Bu¨rger1, Thorsten Mielke1,3, Jan Giesebrecht1,§, Frank Noe´2, Trevor D. Lamb4, Klaus Peter Hofmann1,5, Christian M. T. Spahn1 and Martin Heck1 Research 1Institut fu¨r Medizinische Physik und Biophysik, Charite´ – Universita¨tsmedizin Berlin, corporate member Cite this article: Qureshi BM et al. 2018 It of Freie Universita¨t Berlin, Humboldt-Universita¨t zu Berlin, and Berlin Institute of Health, Berlin, Germany 2Department of Mathematics, Computer Science and Bioinformatics, Freie Universita¨t Berlin, Berlin, Germany takes two transducins to activate the cGMP- 3Microscopy and Cryo Electron Microscopy Group, Max-Planck Institut fu¨r Molekulare Genetik, Berlin, Germany phosphodiesterase 6 in retinal rods. Open Biol. 4Eccles Institute of Neuroscience, John Curtin School of Medical Research, Australian National University, 8: 180075. Canberra, Australian Capital Territory 2600, Australia 5 http://dx.doi.org/10.1098/rsob.180075 Zentrum fu¨r Biophysik und Bioinformatik, Humboldt-Universita¨t zu Berlin, Berlin, Germany BMQ, 0000-0002-0153-7729; TDL, 0000-0003-0299-6115; MH, 0000-0003-0847-5038 Received: 28 April 2018 Accepted: 6 July 2018 Among cyclic nucleotide phosphodiesterases (PDEs), PDE6 is unique in serving as an effector enzyme in G protein-coupled signal transduction. In Subject Area: retinal rods and cones, PDE6 is membrane-bound and activated to hydrolyse biochemistry/biophysics/structural biology its substrate, cGMP, by binding of two active G protein a-subunits (Ga*). To investigate the activation mechanism of mammalian rod PDE6, we Keywords: have collected functional and structural data, and analysed them by reac- PDE6, visual signal transduction, coincidence tion–diffusion simulations. -

Genetic Variation in Phosphodiesterase (PDE) 7B in Chronic Lymphocytic Leukemia: Overview of Genetic Variants of Cyclic Nucleotide Pdes in Human Disease
Journal of Human Genetics (2011) 56, 676–681 & 2011 The Japan Society of Human Genetics All rights reserved 1434-5161/11 $32.00 www.nature.com/jhg ORIGINAL ARTICLE Genetic variation in phosphodiesterase (PDE) 7B in chronic lymphocytic leukemia: overview of genetic variants of cyclic nucleotide PDEs in human disease Ana M Peiro´ 1,2, Chih-Min Tang1, Fiona Murray1,3, Lingzhi Zhang1, Loren M Brown1, Daisy Chou1, Laura Rassenti4, Thomas A Kipps3,4 and Paul A Insel1,3,4 Expression of cyclic adenosine monophosphate-specific phosphodiesterase 7B (PDE7B) mRNA is increased in patients with chronic lymphocytic leukemia (CLL), thus suggesting that variation may occur in the PDE7B gene in CLL. As genetic variation in other PDE family members has been shown to associate with numerous clinical disorders (reviewed in this manuscript), we sought to identify single-nucleotide polymorphisms (SNPs) in the PDE7B gene promoter and coding region of 93 control subjects and 154 CLL patients. We found that the PDE7B gene has a 5¢ non-coding region SNP À347C4T that occurs with similar frequency in CLL patients (1.9%) and controls (2.7%). Tested in vitro, À347C4T has less promoter activity than a wild-type construct. The low frequency of this 5¢ untranslated region variant indicates that it does not explain the higher PDE7B expression in patients with CLL but it has the potential to influence other settings that involve a role for PDE7B. Journal of Human Genetics (2011) 56, 676–681; doi:10.1038/jhg.2011.80; published online 28 July 2011 Keywords: cAMP; chronic lymphocytic -

G Protein-Coupled Receptors
www.aladdin-e.com Address:800 S Wineville Avenue, Ontario, CA 91761,USA Website:www.aladdin-e.com Email USA: [email protected] Email EU: [email protected] Email Asia Pacific: [email protected] G PROTEIN-COUPLED RECEPTORS Overview: The completion of the Human Genome Project allowed the identification of a large family of proteins with a common motif of seven groups of 20–24 hydrophobic amino acids arranged as a-helices. Approximately 800 of these seven transmembrane (7TM) receptors have been identified of which over 300 are non-olfactory receptors (see Fredriksson et al., 2003; Lagerstrom and Schioth, 2008). Subdivision on the basis of sequence homology allows the definition of rhodopsin, secretin, adhesion, glutamate and Frizzled receptor families. NC-IUPHAR recognizes Classes A, B, and C, which equate to the rhodopsin, secretin, and glutamate receptor families. The nomenclature of 7TM receptors is commonly used interchangeably with G protein-coupled receptors (GPCR), although the former nomenclature recognises signalling of 7TM receptors through pathways not involving G proteins. For example, adiponectin and membrane progestin receptors have some sequence homology to 7TM receptors but signal independently of G proteins and appear to reside in membranes in an inverted fashion compared to conventional GPCR. Additionally, the NPR-C natriuretic peptide receptor (see Page S195) has a single transmembrane domain structure, but appears to couple to G proteins to generate cellular responses. The 300+ non-olfactory GPCR are the targets for the majority of drugs in clinical usage (Overington et al., 2006), although only a minority of these receptors are exploited therapeutically. -

G Protein Alpha Inhibitor 1 (GNAI1) Rabbit Polyclonal Antibody Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for TA335036 G protein alpha inhibitor 1 (GNAI1) Rabbit Polyclonal Antibody Product data: Product Type: Primary Antibodies Applications: WB Recommended Dilution: WB Reactivity: Human, Mouse, Rat Host: Rabbit Isotype: IgG Clonality: Polyclonal Immunogen: The immunogen for anti-GNAI1 antibody: synthetic peptide directed towards the middle region of human GNAI1. Synthetic peptide located within the following region: YQLNDSAAYYLNDLDRIAQPNYIPTQQDVLRTRVKTTGIVETHFTFKDLH Formulation: Liquid. Purified antibody supplied in 1x PBS buffer with 0.09% (w/v) sodium azide and 2% sucrose. Note that this product is shipped as lyophilized powder to China customers. Purification: Affinity Purified Conjugation: Unconjugated Storage: Store at -20°C as received. Stability: Stable for 12 months from date of receipt. Predicted Protein Size: 40 kDa Gene Name: G protein subunit alpha i1 Database Link: NP_002060 Entrez Gene 14677 MouseEntrez Gene 25686 RatEntrez Gene 2770 Human P63096 This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 4 G protein alpha inhibitor 1 (GNAI1) Rabbit Polyclonal Antibody – TA335036 Background: Guanine nucleotide-binding proteins (G proteins) form a large family of signal-transducing molecules. They are found as heterotrimers made up of alpha, beta, and gamma subunits. Members of the G protein family have been characterized most extensively on the basis of the alpha subunit, which binds guanine nucleotide, is capable of hydrolyzing GTP, and interacts with specific receptor and effector molecules. -

GNAT1 (NM 144499) Human Tagged ORF Clone Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for RC221246 GNAT1 (NM_144499) Human Tagged ORF Clone Product data: Product Type: Expression Plasmids Product Name: GNAT1 (NM_144499) Human Tagged ORF Clone Tag: Myc-DDK Symbol: GNAT1 Synonyms: CSNB1G; CSNBAD3; GBT1; GNATR Vector: pCMV6-Entry (PS100001) E. coli Selection: Kanamycin (25 ug/mL) Cell Selection: Neomycin ORF Nucleotide >RC221246 representing NM_144499 Sequence: Red=Cloning site Blue=ORF Green=Tags(s) TTTTGTAATACGACTCACTATAGGGCGGCCGGGAATTCGTCGACTGGATCCGGTACCGAGGAGATCTGCC GCCGCGATCGCC ATGGGGGCTGGGGCCAGTGCTGAGGAGAAGCACTCCAGGGAGCTGGAAAAGAAGCTGAAAGAGGACGCTG AGAAGGATGCTCGAACCGTGAAGCTGCTGCTTCTGGGTGCCGGTGAGTCCGGGAAGAGCACCATCGTCAA GCAGATGAAGATTATCCACCAGGACGGGTACTCGCTGGAAGAGTGCCTCGAGTTTATCGCCATCATCTAC GGCAACACGTTGCAGTCCATCCTGGCCATCGTACGCGCCATGACCACACTCAACATCCAGTACGGAGACT CTGCACGCCAGGACGACGCCCGGAAGCTGATGCACATGGCAGACACTATCGAGGAGGGCACGATGCCCAA GGAGATGTCGGACATCATCCAGCGGCTGTGGAAGGACTCCGGTATCCAGGCCTGTTTTGAGCGCGCCTCG GAGTACCAGCTCAACGACTCGGCGGGCTACTACCTCTCCGACCTGGAGCGCCTGGTAACCCCGGGCTACG TGCCCACCGAGCAGGACGTGCTGCGCTCGCGAGTCAAGACCACTGGCATCATCGAGACGCAGTTCTCCTT CAAGGATCTCAACTTCCGGATGTTCGATGTGGGCGGGCAGCGCTCGGAGCGCAAGAAGTGGATCCACTGC TTCGAGGGCGTGACCTGCATCATCTTCATCGCGGCGCTGAGCGCCTACGACATGGTGCTAGTGGAGGACG ACGAAGTGAACCGCATGCACGAGAGCCTGCACCTGTTCAACAGCATCTGCAACCACCGCTACTTCGCCAC GACGTCCATCGTGCTCTTCCTTAACAAGAAGGACGTCTTCTTCGAGAAGATCAAGAAGGCGCACCTCAGC ATCTGTTTCCCGGACTACGATGGACCCAACACCTACGAGGACGCCGGCAACTACATCAAGGTGCAGTTCC