And Mitogen-Activated Protein Kinase (MAPK) Pathways to Enhance Chemoradiotherapy Responsiveness in Colorectal Cancer
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Gross Anatomy
www.BookOfLinks.com THE BIG PICTURE GROSS ANATOMY www.BookOfLinks.com Notice Medicine is an ever-changing science. As new research and clinical experience broaden our knowledge, changes in treatment and drug therapy are required. The authors and the publisher of this work have checked with sources believed to be reliable in their efforts to provide information that is complete and generally in accord with the standards accepted at the time of publication. However, in view of the possibility of human error or changes in medical sciences, neither the authors nor the publisher nor any other party who has been involved in the preparation or publication of this work warrants that the information contained herein is in every respect accurate or complete, and they disclaim all responsibility for any errors or omissions or for the results obtained from use of the information contained in this work. Readers are encouraged to confirm the infor- mation contained herein with other sources. For example and in particular, readers are advised to check the product information sheet included in the package of each drug they plan to administer to be certain that the information contained in this work is accurate and that changes have not been made in the recommended dose or in the contraindications for administration. This recommendation is of particular importance in connection with new or infrequently used drugs. www.BookOfLinks.com THE BIG PICTURE GROSS ANATOMY David A. Morton, PhD Associate Professor Anatomy Director Department of Neurobiology and Anatomy University of Utah School of Medicine Salt Lake City, Utah K. Bo Foreman, PhD, PT Assistant Professor Anatomy Director University of Utah College of Health Salt Lake City, Utah Kurt H. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Normal Gross and Histologic Features of the Gastrointestinal Tract
NORMAL GROSS AND HISTOLOGIC 1 FEATURES OF THE GASTROINTESTINAL TRACT THE NORMAL ESOPHAGUS left gastric, left phrenic, and left hepatic accessory arteries. Veins in the proximal and mid esopha- Anatomy gus drain into the systemic circulation, whereas Gross Anatomy. The adult esophagus is a the short gastric and left gastric veins of the muscular tube measuring approximately 25 cm portal system drain the distal esophagus. Linear and extending from the lower border of the cri- arrays of large caliber veins are unique to the distal coid cartilage to the gastroesophageal junction. esophagus and can be a helpful clue to the site of It lies posterior to the trachea and left atrium a biopsy when extensive cardiac-type mucosa is in the mediastinum but deviates slightly to the present near the gastroesophageal junction (4). left before descending to the diaphragm, where Lymphatic vessels are present in all layers of the it traverses the hiatus and enters the abdomen. esophagus. They drain to paratracheal and deep The subdiaphragmatic esophagus lies against cervical lymph nodes in the cervical esophagus, the posterior surface of the left hepatic lobe (1). bronchial and posterior mediastinal lymph nodes The International Classification of Diseases in the thoracic esophagus, and left gastric lymph and the American Joint Commission on Cancer nodes in the abdominal esophagus. divide the esophagus into upper, middle, and lower thirds, whereas endoscopists measure distance to points in the esophagus relative to the incisors (2). The esophagus begins 15 cm from the incisors and extends 40 cm from the incisors in the average adult (3). The upper and lower esophageal sphincters represent areas of increased resting tone but lack anatomic landmarks; they are located 15 to 18 cm from the incisors and slightly proximal to the gastroesophageal junction, respectively. -

Lorena Novoa-Aponte
A Strain: fl/fl ∆hep Strain: fl/fl ∆hep AnalysisAAV: ofLuc the ironLuc chaperonePCBP1-WT PCBP1PCBP1-∆Fe interactome:PCBP1-∆RNA PCBP1 variant intersection of DNA repair and iron traffickingAAV: Luc Luc WT ∆Fe ∆RNA PCBP1 −40 Lorena Novoa-Aponte a*, Sarju J. Patel a, Olga Protchenko a, James Wohlschlegel b, Caroline C. Philpott a −40 PCBP1, IHC PCBP1, GAPDH - a Genetics and Metabolism Section,a NIDDK, NIH, Bethesda, MD, USA. b Department of Biological Chemistry, UCLA, Los Angeles, CA, USA C.C. Philpott, et al. *[email protected] 80 B P = 0.0262 60 40 1. Iron is essential but toxic 2. Increased DNA damage in cells lacking PCBP1 ns H&E ? + 20 C.C. Philpott, et al. Iron is used as an essential cofactor by several enzymes involved in DNA ⦿ Increased TUNEL in cells and tissue livers from mice lacking PCBP1. ? replication and repair. However, unchaperoned iron promotes redox ⦿ PCBP1 binds both, iron and single-stranded nucleicTG, nmol/mg protein 0 acids. stress that may affect DNA stability. Strain: Δhep Δhep Δhep ? ⦿ The iron binding activity of PCBP1 controls suppressionAAV: of DNA damage C WT ∆Fe ∆RNA PCBP1 variant ? Iron storage HEK293 cells Mouse Liver ? A 8 siNT+P1var 100 ? A 8 var siPCBP1 2 ? Mammals use the iron siNT+P1 ns var Fe-S cluster assembly 6 siPCBP1+P1siPCBP1 80 ADI1 TUNEL chaperone PCBP1 to var ? 6 siPCBP1+P1 metalate several iron- 60 = 0.0468 = 0.0354 = 0.0465 cell / mm population P P P population + dependent enzymes + 4 + 4 ? 40 Degradation of HIF1α Fig. 3. Iron chaperone-mediated handling of cytosolic labile iron pool. -

Whole Egg Consumption Increases Gene Expression Within the Glutathione Pathway in the Liver of Zucker Diabetic Fatty Rats
Food Science and Human Nutrition Publications Food Science and Human Nutrition 11-3-2020 Whole egg consumption increases gene expression within the glutathione pathway in the liver of Zucker Diabetic Fatty rats Joe L. Webb Iowa State University Amanda E. Bries Iowa State University, [email protected] Brooke Vogel Iowa State University Claudia Carrillo Iowa State University, [email protected] Lily Harvison Iowa State University, [email protected] See next page for additional authors Follow this and additional works at: https://lib.dr.iastate.edu/fshn_hs_pubs Part of the Dietetics and Clinical Nutrition Commons, Endocrinology, Diabetes, and Metabolism Commons, Exercise Science Commons, Food Chemistry Commons, Human and Clinical Nutrition Commons, and the Molecular, Genetic, and Biochemical Nutrition Commons The complete bibliographic information for this item can be found at https://lib.dr.iastate.edu/ fshn_hs_pubs/38. For information on how to cite this item, please visit http://lib.dr.iastate.edu/ howtocite.html. This Article is brought to you for free and open access by the Food Science and Human Nutrition at Iowa State University Digital Repository. It has been accepted for inclusion in Food Science and Human Nutrition Publications by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. Whole egg consumption increases gene expression within the glutathione pathway in the liver of Zucker Diabetic Fatty rats Abstract Nutrigenomic evidence supports the idea that Type 2 Diabetes Mellitus (T2DM) arises due to the interactions between the transcriptome, individual genetic profiles, lifestyle, and diet. Since eggs are a nutrient dense food containing bioactive ingredients that modify gene expression, our goal was to examine the role of whole egg consumption on the transcriptome during T2DM. -
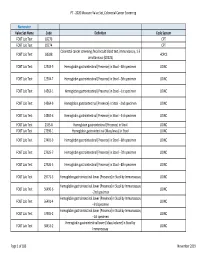
2020 Measure Value Set Colorectal Cancer Screening
PT ‐ 2020 Measure Value Set_Colorectal Cancer Screening Numerator Value Set Name Code Definition Code System FOBT Lab Test 82270 CPT FOBT Lab Test 82274 CPT Colorectal cancer screening; fecal occult blood test, immunoassay, 1‐3 FOBT Lab Test G0328 HCPCS simultaneous (G0328) FOBT Lab Test 12503‐9 Hemoglobin.gastrointestinal [Presence] in Stool ‐‐4th specimen LOINC FOBT Lab Test 12504‐7 Hemoglobin.gastrointestinal [Presence] in Stool ‐‐5th specimen LOINC FOBT Lab Test 14563‐1 Hemoglobin.gastrointestinal [Presence] in Stool ‐‐1st specimen LOINC FOBT Lab Test 14564‐9 Hemoglobin.gastrointestinal [Presence] in Stool ‐‐2nd specimen LOINC FOBT Lab Test 14565‐6 Hemoglobin.gastrointestinal [Presence] in Stool ‐‐3rd specimen LOINC FOBT Lab Test 2335‐8 Hemoglobin.gastrointestinal [Presence] in Stool LOINC FOBT Lab Test 27396‐1 Hemoglobin.gastrointestinal [Mass/mass] in Stool LOINC FOBT Lab Test 27401‐9 Hemoglobin.gastrointestinal [Presence] in Stool ‐‐6th specimen LOINC FOBT Lab Test 27925‐7 Hemoglobin.gastrointestinal [Presence] in Stool ‐‐7th specimen LOINC FOBT Lab Test 27926‐5 Hemoglobin.gastrointestinal [Presence] in Stool ‐‐8th specimen LOINC FOBT Lab Test 29771‐3 Hemoglobin.gastrointestinal.lower [Presence] in Stool by Immunoassay LOINC Hemoglobin.gastrointestinal.lower [Presence] in Stool by Immunoassay FOBT Lab Test 56490‐6 LOINC ‐‐2nd specimen Hemoglobin.gastrointestinal.lower [Presence] in Stool by Immunoassay FOBT Lab Test 56491‐4 LOINC ‐‐3rd specimen Hemoglobin.gastrointestinal.lower [Presence] in Stool by Immunoassay FOBT Lab Test 57905‐2 -

Qt38n028mr Nosplash A3e1d84
! ""! ACKNOWLEDGEMENTS I dedicate this thesis to my parents who inspired me to become a scientist through invigorating scientific discussions at the dinner table even when I was too young to understand what the hippocampus was. They also prepared me for the ups and downs of science and supported me through all of these experiences. I would like to thank my advisor Dr. Elizabeth Blackburn and my thesis committee members Dr. Eric Verdin, and Dr. Emmanuelle Passegue. Liz created a nurturing and supportive environment for me to explore my own ideas, while at the same time teaching me how to love science, test my questions, and of course provide endless ways to think about telomeres and telomerase. Eric and Emmanuelle both gave specific critical advice about the proper experiments for T cells and both volunteered their lab members for further critical advice. I always felt inspired with a sense of direction after thesis committee meetings. The Blackburn lab is full of smart and dedicated scientists whom I am thankful for their support. Specifically Dr. Shang Li and Dr. Brad Stohr for their stimulating scientific debates and “arguments.” Dr. Jue Lin, Dana Smith, Kyle Lapham, Dr. Tet Matsuguchi, and Kyle Jay for their friendships and discussions about what my data could possibly mean. Dr. Eva Samal for teaching me molecular biology techniques and putting up with my late night lab exercises. Beth Cimini for her expertise with microscopy, FACs, singing, and most of all for being a caring and supportive friend. Finally, I would like to thank Dr. Imke Listerman, my scientific partner for most of the breast cancer experiments. -

Thieme: Lymphedema Management
8 1 Anatomy tween a proximal and a distal pair of valves is called lymph angion (Fig. 1−4). The media in valvular areas of lymph collectors contains less smooth musculature than the angion area. Lymph angions have an autonomic contraction frequency of ෂ10 to 12 contractions per minute at rest (lymphangiomotoricity). In healthy lymph collectors, the proximal valve is open during the systole, whereas the distal valve is closed; in the diastole, the op- posite is the case. This permits directional flow of lymph fluid from distal to proximal angions. In lymphangiectasia (dilation) with valvular insufficiency, the lymph flow may reverse into distal lymph angions (lymphatic reflux). Lymph collectors have the ability to react to an increase in lymph formation with an in- crease in contraction frequency. The increase in lymph fluid entering the lymph angion will cause a stretch on the wall of the angion, which Figure 1−4 Lymph collectors. 1. Lymph collector; 2. Afferent lymph collector to lymph node; 3. Efferent in turn results in an increase in lymphangio- lymph collector from lymph node; 4. Lymph node; 5. motoricity (lymphatic safety factor; see also Cross section through a lymph collector in the area of Chapter 2, Safety Factor of the Lymphatic Sys- the valves; 6. Lymph angion. tem). Other factors that may influence lymphan- giomotoricity are external stretch on the It is postulated that the main purpose of pre- lymph angion wall (e.g., manual lymph collectors is the transport of lymph fluid from drainage), temperature, activity of muscle and the capillaries to lymph collectors. Due to the joint pumps, diaphragmatic breathing, pulsa- capillary-like wall structure in some areas, pre- tion of adjacent arteries, and certain tissue collectors are able to absorb lymphatic loads. -

The Inverse Agonist DG172 Triggers a Pparβ/Δ-Independent Myeloid Lineage Shift and Promotes GM-CSF/IL-4-Induced Dendritic Cell Differentiation
Downloaded from molpharm.aspetjournals.org at ASPET Journals on September 26, 2021 1 MOL #94672 #94672 MOL -independent myeloid lineage shift and -independent myeloid lineage δ / β nagel, Wolfgang Meissner, Gavin Giehl, Cornelia Giehl, Cornelia Gavin Wolfgang Meissner, nagel, This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. This article has not been copyedited and formatted. The final version may differ from this version. -

Hypermutation of DPYD Deregulates Pyrimidine Metabolism and Promotes Malignant Progression Lauren Edwards, Rohit Gupta, and Fabian Volker Filipp
Published OnlineFirst November 25, 2015; DOI: 10.1158/1541-7786.MCR-15-0403 Genomics Molecular Cancer Research Hypermutation of DPYD Deregulates Pyrimidine Metabolism and Promotes Malignant Progression Lauren Edwards, Rohit Gupta, and Fabian Volker Filipp Abstract New strategies are needed to diagnose and target human the background mutation rate. Structural analysis of the DPYD melanoma. To this end, genomic analyses was performed to protein dimer reveals a potential hotspot of recurring somatic assess somatic mutations and gene expression signatures using mutations in the ligand-binding sites as well as the interfaces of a large cohort of human skin cutaneous melanoma (SKCM) protein domains that mediated electron transfer. Somatic muta- patients from The Cancer Genome Atlas (TCGA) project to tions of DPYD are associated with upregulation of pyrimidine identify critical differences between primary and metastatic degradation, nucleotide synthesis, and nucleic acid processing tumors. Interestingly, pyrimidine metabolism is one of the major while salvage and nucleotide conversion is downregulated in pathways to be significantly enriched and deregulated at the TCGA SKCM. transcriptional level in melanoma progression. In addition, dihy- dropyrimidine dehydrogenase (DPYD) and other important Implications: At a systems biology level, somatic mutations of pyrimidine-related genes: DPYS, AK9, CAD, CANT1, ENTPD1, DPYD cause a switch in pyrimidine metabolism and promote NME6, NT5C1A, POLE, POLQ, POLR3B, PRIM2, REV3L, and gene expression of pyrimidine enzymes toward malignant pro- UPP2 are significantly enriched in somatic mutations relative to gression. Mol Cancer Res; 14(2); 196–206. Ó2015 AACR. Introduction Pyrimidine synthesis is a key metabolic bottleneck important for DNA replication in tumor cells and, therefore, represents a Cancer cells take advantage of distinct metabolic pathways valuable diagnostic and therapeutic target. -

Investigating the Impact of Telomere Dysfunction on the Chronic Lymphocytic Leukaemia Genome
INVESTIGATING THE IMPACT OF TELOMERE DYSFUNCTION ON THE CHRONIC LYMPHOCYTIC LEUKAEMIA GENOME Laura Escudero-Monreal A thesis submitted for the degree of Doctor of Philosophy September 2017 Institute of Cancer and Genetics School of Medicine Cardiff University Funded by Cancer Research Wales and Cardiff University I dedicate this Ph.D. thesis to everyone who suffers from cancer, for I hope one day, science can fight and win this battle. Laura Escudero-Monreal NOTICE OF SUBMISSION DECLARATION This work has not been submitted in substance for any other degree or award at this or any other university or place of learning, nor is being submitted concurrently in candidature for any degree or other award. Signed: Date: STATEMENT 1 This thesis is being submitted in partial fulfilment of the requirements for the degree of PhD. Signed: Date: STATEMENT 2 This thesis is the result of my own independent work/investigation, except where otherwise stated, and the thesis has not been edited by a third party beyond what is permitted by Cardiff University’s Policy on the Use of Third Party Editors by Research Degree Students. Other sources are acknowledged by explicit references. The views expressed are my own. Signed: Date: STATEMENT 3 I hereby give consent for my thesis, if accepted, to be available online in the University’s Open Access repository and for inter-library loan, and for the title and summary to be made available to outside organisations. Signed: Date: STATEMENT 4: PREVIOUSLY APPROVED BAR ON ACCESS I hereby give consent for my thesis, if accepted, to be available online in the University’s Open Access repository and for inter-library loans after expiry of a bar on access previously approved by the Academic Standards & Quality Committee. -

Thesis Template
Characterisation of the Co-chaperone Small Glutamine-rich Tetratricopeptide Repeat containing protein alpha as a Regulator of Androgen Receptor Activity in Prostate Cancer Cells A thesis submitted to the University of Adelaide in total fulfilment of the requirements for the degree of Doctor of Philosophy by ANDREW PAUL TROTTA B.Sc. (Mol. Biol.), B.Sc. (Hons) Department of Medicine The University of Adelaide Adelaide, South Australia July 2011 This thesis is dedicated to my mum and dad. Thank you for all your love and support. DECLARATION I ACKNOWLEDGEMENTS II ABBREVIATIONS V ABSTRACT X CHAPTER 1: INTRODUCTION 2 1.1 Overview 2 1.2 Development of the prostate 4 1.2.1 Androgen physiology 4 1.2.2 Development of the normal prostate 5 1.3 Prostate cancer and progression 9 1.3.1 Pathogenesis 9 1.4 Diagnosis 10 1.4.1 Clinically localized and advanced disease 11 1.5 Treatment 12 1.5.1 Localised Disease 12 1.5.2 Metastatic Disease 12 1.6 The androgen signalling axis 14 1.6.1 The androgen receptor 15 1.6.2 The androgen receptor gene 16 1.6.3 The androgen receptor protein and domains 19 1.7 Androgen receptor co-regulators 24 1.7.1 Co-activators 24 1.7.2 Co-repressors 25 1.7.3 Chaperones 26 1.8 The molecular chaperone complex and androgen receptor maturation 27 1.8.1 Chaperones involved in ligand binding and nuclear translocation 32 1.8.2 Chaperones and transcriptional activation 37 1.9 Chaperones in prostate cancer 37 1.10 Chaperones as therapeutic targets 39 1.11 Tetratricopeptide repeat containing co-chaperones 40 1.11.1 Structure of TPR domain