Request for Applications (RFA)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
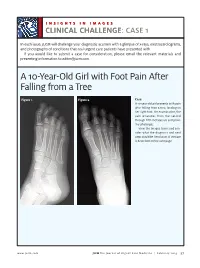
A 10-Year-Old Girl with Foot Pain After Falling from a Tree
INSIGHTS IN IMAGES CLINICAL CHALLENGECHALLENGE: CASE 1 In each issue, JUCM will challenge your diagnostic acumen with a glimpse of x-rays, electrocardiograms, and photographs of conditions that real urgent care patients have presented with. If you would like to submit a case for consideration, please email the relevant materials and presenting information to [email protected]. A 10-Year-Old Girl with Foot Pain After Falling from a Tree Figure 1. Figure 2. Case A 10-year-old girl presents with pain after falling from a tree, landing on her right foot. On examination, the pain emanates from the second through fifth metatarsals and proxi- mal phalanges. View the images taken and con- sider what the diagnosis and next steps would be. Resolution of the case is described on the next page. www.jucm.com JUCM The Journal of Urgent Care Medicine | February 2019 37 INSIGHTS IN IMAGES: CLINICAL CHALLENGE THE RESOLUTION Figure 1. Mediastinal air Figure 1. Differential Diagnosis Pearls for Urgent Care Management and Ⅲ Fracture of the distal fourth metatarsal Considerations for Transfer Ⅲ Plantar plate disruption Ⅲ Emergent transfer should be considered with associated neu- Ⅲ Sesamoiditis rologic deficit, compartment syndrome, open fracture, or vas- Ⅲ Turf toe cular compromise Ⅲ Referral to an orthopedist is warranted in the case of an in- Diagnosis tra-articular fracture, or with Lisfranc ligament injury or ten- Angulation of the distal fourth metatarsal metaphyseal cortex derness over the Lisfranc ligament and hairline lucency consistent with fracture. Acknowledgment: Images courtesy of Teleradiology Associates. Learnings/What to Look for Ⅲ Proximal metatarsal fractures are most often caused by crush- ing or direct blows Ⅲ In athletes, an axial load placed on a plantar-flexed foot should raise suspicion of a Lisfranc injury 38 JUCM The Journal of Urgent Care Medicine | February 2019 www.jucm.com INSIGHTS IN IMAGES CLINICAL CHALLENGE: CASE 2 A 55-Year-Old Man with 3 Hours of Epigastric Pain 55 years PR 249 QRSD 90 QT 471 QTc 425 AXES P 64 QRS -35 T 30 Figure 1. -

Genetic Mechanisms of Disease in Children: a New Look
Genetic Mechanisms of Disease in Children: A New Look Laurie Demmer, MD Tufts Medical Center and the Floating Hospital for Children Boston, MA Traditionally genetic disorders have been linked to the ‘one gene-one protein-one disease’ hypothesis. However recent advances in the field of molecular biology and biotechnology have afforded us the opportunity to greatly expand our knowledge of genetics, and we now know that the mechanisms of inherited disorders are often significantly more complex, and consequently, much more intriguing, than originally thought. Classical mendelian disorders with relatively simple genetic mechanisms do exist, but turn out to be far more rare than originally thought. All patients with sickle cell disease for example, carry the same A-to-T point mutation in the sixth codon of the beta globin gene. This results in a glutamate to valine substitution which changes the shape and the function of the globin molecule in a predictable way. Similarly, all patients with achondroplasia have a single base pair substitution at nucleotide #1138 of the FGFR3 gene. On the other hand, another common inherited disorder, cystic fibrosis, is known to result from changes in a specific transmembrane receptor (CFTR), but over 1000 different disease-causing mutations have been reported in this single gene. Since most commercial labs only test for between 23-100 different mutations, interpreting CFTR mutation testing is significantly complicated by the known risk of false negative results. Many examples of complex, or non-Mendelian, inheritance are now known to exist and include disorders of trinucleotide repeats, errors in imprinting, and gene dosage effects. -

Segmental Overvekst Og Vaskulærmalformasjoner V02
2/1/2021 Segmental overvekst og vaskulærmalformasjoner v02 Avdeling for medisinsk genetikk Segmental overvekst og vaskulærmalformasjoner Genpanel, versjon v02 * Enkelte genomiske regioner har lav eller ingen sekvensdekning ved eksomsekvensering. Dette skyldes at de har stor likhet med andre områder i genomet, slik at spesifikk gjenkjennelse av disse områdene og påvisning av varianter i disse områdene, blir vanskelig og upålitelig. Disse genetiske regionene har vi identifisert ved å benytte USCS segmental duplication hvor områder større enn 1 kb og ≥90% likhet med andre regioner i genomet, gjenkjennes (https://genome.ucsc.edu). Vi gjør oppmerksom på at ved identifiseringav ekson oppstrøms for startkodon kan eksonnummereringen endres uten at transkript ID endres. Avdelingens websider har en full oversikt over områder som er affisert av segmentale duplikasjoner. ** Transkriptets kodende ekson. Ekson Gen Gen affisert (HGNC (HGNC Transkript Ekson** Fenotype av symbol) ID) segdup* ACVRL1 175 NM_000020.3 2-10 Telangiectasia, hereditary hemorrhagic, type 2 OMIM ADAMTS3 219 NM_014243.3 1-22 Hennekam lymphangiectasia- lymphedema syndrome 3 OMIM AKT1 391 NM_005163.2 2-14 Cowden syndrome 6 OMIM Proteus syndrome, somatic OMIM AKT2 392 NM_001626.6 2-14 Diabetes mellitus, type II OMIM Hypoinsulinemic hypoglycemia with hemihypertrophy OMIM AKT3 393 NM_005465.7 2-14 Megalencephaly-polymicrogyria- polydactyly-hydrocephalus syndrome 2 OMIM file:///data/SegOv_v02-web.html 1/7 2/1/2021 Segmental overvekst og vaskulærmalformasjoner v02 Ekson Gen Gen affisert (HGNC (HGNC -

My Beloved Neutrophil Dr Boxer 2014 Neutropenia Family Conference
The Beloved Neutrophil: Its Function in Health and Disease Stem Cell Multipotent Progenitor Myeloid Lymphoid CMP IL-3, SCF, GM-CSF CLP Committed Progenitor MEP GMP GM-CSF, IL-3, SCF EPO TPO G-CSF M-CSF IL-5 IL-3 SCF RBC Platelet Neutrophil Monocyte/ Basophil B-cells Macrophage Eosinophil T-Cells Mast cell NK cells Mature Cell Dendritic cells PRODUCTION AND KINETICS OF NEUTROPHILS CELLS % CELLS TIME Bone Marrow: Myeloblast 1 7 - 9 Mitotic Promyelocyte 4 Days Myelocyte 16 Maturation/ Metamyelocyte 22 3 – 7 Storage Band 30 Days Seg 21 Vascular: Peripheral Blood Seg 2 6 – 12 hours 3 Marginating Pool Apoptosis and ? Tissue clearance by 0 – 3 macrophages days PHAGOCYTOSIS 1. Mobilization 2. Chemotaxis 3. Recognition (Opsonization) 4. Ingestion 5. Degranulation 6. Peroxidation 7. Killing and Digestion 8. Net formation Adhesion: β 2 Integrins ▪ Heterodimer of a and b chain ▪ Tight adhesion, migration, ingestion, co- stimulation of other PMN responses LFA-1 Mac-1 (CR3) p150,95 a2b2 a CD11a CD11b CD11c CD11d b CD18 CD18 CD18 CD18 Cells All PMN, Dendritic Mac, mono, leukocytes mono/mac, PMN, T cell LGL Ligands ICAMs ICAM-1 C3bi, ICAM-3, C3bi other other Fibrinogen other GRANULOCYTE CHEMOATTRACTANTS Chemoattractants Source Activators Lipids PAF Neutrophils C5a, LPS, FMLP Endothelium LTB4 Neutrophils FMLP, C5a, LPS Chemokines (a) IL-8 Monocytes, endothelium LPS, IL-1, TNF, IL-3 other cells Gro a, b, g Monocytes, endothelium IL-1, TNF other cells NAP-2 Activated platelets Platelet activation Others FMLP Bacteria C5a Activation of complement Other Important Receptors on PMNs ñ Pattern recognition receptors – Detect microbes - Toll receptor family - Mannose receptor - bGlucan receptor – fungal cell walls ñ Cytokine receptors – enhance PMN function - G-CSF, GM-CSF - TNF Receptor ñ Opsonin receptors – trigger phagocytosis - FcgRI, II, III - Complement receptors – ñ Mac1/CR3 (CD11b/CD18) – C3bi ñ CR-1 – C3b, C4b, C3bi, C1q, Mannose binding protein From JG Hirsch, J Exp Med 116:827, 1962, with permission. -

Megalencephaly and Macrocephaly
277 Megalencephaly and Macrocephaly KellenD.Winden,MD,PhD1 Christopher J. Yuskaitis, MD, PhD1 Annapurna Poduri, MD, MPH2 1 Department of Neurology, Boston Children’s Hospital, Boston, Address for correspondence Annapurna Poduri, Epilepsy Genetics Massachusetts Program, Division of Epilepsy and Clinical Electrophysiology, 2 Epilepsy Genetics Program, Division of Epilepsy and Clinical Department of Neurology, Fegan 9, Boston Children’s Hospital, 300 Electrophysiology, Department of Neurology, Boston Children’s Longwood Avenue, Boston, MA 02115 Hospital, Boston, Massachusetts (e-mail: [email protected]). Semin Neurol 2015;35:277–287. Abstract Megalencephaly is a developmental disorder characterized by brain overgrowth secondary to increased size and/or numbers of neurons and glia. These disorders can be divided into metabolic and developmental categories based on their molecular etiologies. Metabolic megalencephalies are mostly caused by genetic defects in cellular metabolism, whereas developmental megalencephalies have recently been shown to be caused by alterations in signaling pathways that regulate neuronal replication, growth, and migration. These disorders often lead to epilepsy, developmental disabilities, and Keywords behavioral problems; specific disorders have associations with overgrowth or abnor- ► megalencephaly malities in other tissues. The molecular underpinnings of many of these disorders are ► hemimegalencephaly now understood, providing insight into how dysregulation of critical pathways leads to ► -

MECHANISMS in ENDOCRINOLOGY: Novel Genetic Causes of Short Stature
J M Wit and others Genetics of short stature 174:4 R145–R173 Review MECHANISMS IN ENDOCRINOLOGY Novel genetic causes of short stature 1 1 2 2 Jan M Wit , Wilma Oostdijk , Monique Losekoot , Hermine A van Duyvenvoorde , Correspondence Claudia A L Ruivenkamp2 and Sarina G Kant2 should be addressed to J M Wit Departments of 1Paediatrics and 2Clinical Genetics, Leiden University Medical Center, PO Box 9600, 2300 RC Leiden, Email The Netherlands [email protected] Abstract The fast technological development, particularly single nucleotide polymorphism array, array-comparative genomic hybridization, and whole exome sequencing, has led to the discovery of many novel genetic causes of growth failure. In this review we discuss a selection of these, according to a diagnostic classification centred on the epiphyseal growth plate. We successively discuss disorders in hormone signalling, paracrine factors, matrix molecules, intracellular pathways, and fundamental cellular processes, followed by chromosomal aberrations including copy number variants (CNVs) and imprinting disorders associated with short stature. Many novel causes of GH deficiency (GHD) as part of combined pituitary hormone deficiency have been uncovered. The most frequent genetic causes of isolated GHD are GH1 and GHRHR defects, but several novel causes have recently been found, such as GHSR, RNPC3, and IFT172 mutations. Besides well-defined causes of GH insensitivity (GHR, STAT5B, IGFALS, IGF1 defects), disorders of NFkB signalling, STAT3 and IGF2 have recently been discovered. Heterozygous IGF1R defects are a relatively frequent cause of prenatal and postnatal growth retardation. TRHA mutations cause a syndromic form of short stature with elevated T3/T4 ratio. Disorders of signalling of various paracrine factors (FGFs, BMPs, WNTs, PTHrP/IHH, and CNP/NPR2) or genetic defects affecting cartilage extracellular matrix usually cause disproportionate short stature. -

Costello Syndrome
orphananesthesia Anaesthesia recommendations for patients suffering from Costello syndrome Disease name: Costello syndrome ICD 10: Q87.8 Synonyms: Significant phenotypical overlap with CFC (cardiofaciocutaneous syndrome) and Noonan syndrome. Disease summary: Costello syndrome (CS) is a rare disorder (so-called RAS-opathy, see below), affecting up to 300 people worldwide. First described by Dr Jack Costello in 1977, the syndrome is characterised by failure to thrive (FTT), poor feeding, short stature, developmental delay, distinctive facial features, excessive loose skin, cardiac abnormalities, and an increased risk of tumour development. RAS is a family of genes coding for small GTPases and includes amongst others HRAS. The HRAS gene is a proto-oncogene, which forms part of the MAPK (mitogen activated protein kinase) signalling pathway. Up-regulation of this signalling pathway causes unopposed cell growth, causing tumour predisposition. The MAPK pathway is also the site of mutations causing both CFC and Noonan syndrome. CS can be caused by a number of mutations in the HRAS gene. Most mutations do occur de novo, but there is some evidence that a minority are inherited in an autosomal dominant manner. Patients with CS are born large for gestational age, and there is a strong association with polyhydramnios and preterm labour. Growth later slows due to feeding difficulties. Head circumference is affected to a lesser degree than height and weight, which gives rise to relative macrocephaly. Growth Hormone (GH) deficiency can cause neonatal hypoglycaemia, and contributes to growth retardation. The disease is characterised by distinctive facial features including downslanting palpebral fissures, epicanthic folds, ptosis, flattened nasal bridge (hypertelorism), low set ears, thick lips, macroglossia and short neck. -

Workshop Schedule
Workshop Schedule Friday, August 27th Registration: 4:00‐6:30 pm Dinner 6:30‐8:00 pm Reception 8:00‐10:00 Saturday, August 28th 7:00‐8:15 am Breakfast: 8:15am Welcome and Introductions 8:30‐10:15 am Session 1: Novel strategies to understand the causes/mechanisms of birth defects (Moderator: Angela Lin) 8:30 am Les Biesecker ‐ Using massively parallel sequencing technologies to provide improved molecular delineation of human malformation syndromes 9:15 am Bamshad ‐ Exome sequencing identifies a gene for Kabuki syndrome 9:30 am Boycott ‐ Next‐Generation sequencing strategies give insight into the mechanism of birth defects in a Canadian isolated population 9:45 am Bleyl ‐ Comparison of pooled allelic ratios (CoPAR) analysis: an efficient method for mapping genetic traits in extended pedigrees 10:00 am Allanson – Nablus mask‐like facial syndrome and blepharo‐naso‐facial syndrome are the same entity. Refinement of the critical region of chromosome 8q22.1 points to a potential candidate gene 10:15‐10:45 am BREAK 10:45‐12:00 pm Session 2: Novel strategies to understand the causes/mechanisms of birth defects (cont.) (Moderator: Mike Bamshad) 10:45 am Krantz ‐ Applying novel genomic tools towards understanding an old chromosomal diagnosis: Using genome‐wide expression and SNP genotyping to identify the true cause of Pallister‐Killian syndrome 11:00 am Paciorkowski ‐ Bioinformatic analysis of published and novel copy number variants suggests candidate genes and networks for infantile spasms 11:15 am Bernier ‐ Identification of a novel Fibulin -

Blueprint Genetics Bone Marrow Failure Syndrome Panel
Bone Marrow Failure Syndrome Panel Test code: HE0801 Is a 135 gene panel that includes assessment of non-coding variants. Is ideal for patients with a clinical suspicion of inherited bone marrow failure syndromes. The genes on this panel are included in the Comprehensive Hematology Panel. About Bone Marrow Failure Syndrome Inherited bone marrow failure syndromes (IBMFS) are a diverse set of genetic disorders characterized by the inability of the bone marrow to produce sufficient circulating blood cells. Bone marrow failure can affect all blood cell lineages causing clinical symptoms similar to aplastic anemia, or be restricted to one or two blood cell lineages. The clinical presentation may include thrombocytopenia or neutropenia. Hematological manifestations may be accompanied by physical features such as short stature and abnormal skin pigmentation in Fanconi anemia and dystrophic nails, lacy reticular pigmentation and oral leukoplakia in dyskeratosis congenita. Patients with IBMFS have an increased risk of developing cancer—either hematological or solid tumors. Early and correct disease recognition is important for management and surveillance of the diseases. Currently, accurate genetic diagnosis is essential to confirm the clinical diagnosis. The most common phenotypes that are covered by the panel are Fanconi anemia, Diamond-Blackfan anemia, dyskeratosis congenita, Shwachman-Diamond syndrome and WAS-related disorders. Availability 4 weeks Gene Set Description Genes in the Bone Marrow Failure Syndrome Panel and their clinical significance -

Noonan Spectrum Disorders Panel, Sequencing ARUP Test Code 2010772 Noonan Disorders Sequencing Specimen Whole Blood
Patient Report |FINAL Client: Example Client ABC123 Patient: Patient, Example 123 Test Drive Salt Lake City, UT 84108 DOB 12/9/2011 UNITED STATES Gender: Female Patient Identifiers: 01234567890ABCD, 012345 Physician: Doctor, Example Visit Number (FIN): 01234567890ABCD Collection Date: 01/01/2017 12:34 Noonan Spectrum Disorders Panel, Sequencing ARUP test code 2010772 Noonan Disorders Sequencing Specimen Whole Blood Noonan Disorders Sequencing Interp Positive INDICATION FOR TESTING Short stature and facial features suggestive of Noonan syndrome. RESULT One pathogenic variant was detected in the PTPN11 gene. PATHOGENIC VARIANT Gene: PTPN11 (NM_002834.3) Nucleic Acid Change: c.188A>G; Heterozygous Amino Acid Alteration: p.Tyr63Cys Inheritance: Autosomal Dominant INTERPRETATION One pathogenic variant, c.188A>G; p.Tyr63Cys, was detected in the PTPN11 gene by massively parallel sequencing and confirmed by Sanger sequencing. Pathogenic PTPN11 variants are inherited in an autosomal dominant manner, and are associated with Noonan syndrome 1 (MIM: 163950), metachondromatosis (MIM: 156250), and LEOPARD syndrome 1 (MIM: 151100). No additional pathogenic variants were identified in the other targeted genes by massively parallel sequencing. Please refer to the background information included in this report for a list of the genes analyzed and limitations of this test. Evidence for variant classification: The PTPN11 c.188A>G, p.Tyr63Cys variant (rs121918459) has been reported in multiple patients diagnosed with Noonan syndrome (Tartaglia 2001, Jongmans 2011, Martinelli 2012, Hashida 2013, Lepri 2014, Okamoto 2015). This variant is located in a structurally important region of the catalytic N-terminal SH2 domain of PTPN11 (Hof 1998), and several additional variants in neighboring codons have also been identified in Noonan patients (Jongmans 2011, Tartaglia 2002, Tartaglia 2006). -

Costello Syndrome with Severe Nodulocystic Acne: Unexpected Significant Improvement of Acanthosis Nigricans After Oral Isotretinoin Treatment
Hindawi Publishing Corporation Case Reports in Pediatrics Volume 2015, Article ID 934865, 4 pages http://dx.doi.org/10.1155/2015/934865 Case Report Costello Syndrome with Severe Nodulocystic Acne: Unexpected Significant Improvement of Acanthosis Nigricans after Oral Isotretinoin Treatment Leelawadee Sriboonnark,1 Harleen Arora,2 Leyre Falto-Aizpurua,2 Sonal Choudhary,2 and Elizabeth Alvarez Connelly2 1 Division of Dermatology, Department of Pediatrics, Faculty of Medicine, Khon Kaen University, Khon Kaen 40002, Thailand 2Miller School of Medicine, Department of Pediatric Dermatology, University of Miami, 1600 NW 10th Avenue, Rosenstiel Medical Science Building, Room 2023, Miami, FL 33136, USA Correspondence should be addressed to Leelawadee Sriboonnark; [email protected] Received 1 November 2014; Accepted 23 February 2015 Academic Editor: Ozgur Cogulu Copyright © 2015 Leelawadee Sriboonnark et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. We report the case of 17-year-old female diagnosed with Costello syndrome. Genetic testing provided a proof with G12S mutation in the HRAS gene since 3 years of age with a presentation of severe nodulocystic acne on her face. After 2 months of oral isotretinoin treatment, improvement in her acne was observed. Interestingly, an unexpected significant improvement of acanthosis nigricans on her neck and dorsum of her hands was found as well. We present this case as a successful treatment option by using oral isotretinoin for the treatment of acanthosis nigricans in Costello syndrome patients. 1. Introduction revealedamutation(G12S)intheHRASgene.Shealso has typical features of this syndrome which included coarse Costello syndrome is an autosomal dominant inherited facies (Figure 1), sparse hair, full lips, redundant skin, ulnar disorder with frequent de novo mutation in the HRAS deviation of the wrists and fingers, developmental delay, and gene [1]. -

Essential Genetics 5
Essential genetics 5 Disease map on chromosomes 例 Gaucher disease 単一遺伝子病 天使病院 Prader-Willi syndrome 隣接遺伝子症候群,欠失が主因となる疾患 臨床遺伝診療室 外木秀文 Trisomy 13 複数の遺伝子の重複によって起こる疾患 挿画 Koromo 遺伝子の座位あるいは欠失等の範囲を示す Copyright (c) 2010 Social Medical Corporation BOKOI All Rights Reserved. Disease map on chromosome 1 Gaucher disease Chromosome 1q21.1 1p36 deletion syndrome deletion syndrome Adrenoleukodystrophy, neonatal Cardiomyopathy, dilated, 1A Zellweger syndrome Charcot-Marie-Tooth disease Emery-Dreifuss muscular Hypercholesterolemia, familial dystrophy Hutchinson-Gilford progeria Ehlers-Danlos syndrome, type VI Muscular dystrophy, limb-girdle type Congenital disorder of Insensitivity to pain, congenital, glycosylation, type Ic with anhidrosis Diamond-Blackfan anemia 6 Charcot-Marie-Tooth disease Dejerine-Sottas syndrome Marshall syndrome Stickler syndrome, type II Chronic granulomatous disease due to deficiency of NCF-2 Alagille syndrome 2 Copyright (c) 2010 Social Medical Corporation BOKOI All Rights Reserved. Disease map on chromosome 2 Epiphyseal dysplasia, multiple Spondyloepimetaphyseal dysplasia Brachydactyly, type D-E, Noonan syndrome Brachydactyly-syndactyly syndrome Peters anomaly Synpolydactyly, type II and V Parkinson disease, familial Leigh syndrome Seizures, benign familial Multiple pterygium syndrome neonatal-infantile Escobar syndrome Ehlers-Danlos syndrome, Brachydactyly, type A1 type I, III, IV Waardenburg syndrome Rhizomelic chondrodysplasia punctata, type 3 Alport syndrome, autosomal recessive Split-hand/foot malformation Crigler-Najjar