Blueprint Genetics Noonan Syndrome Panel
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
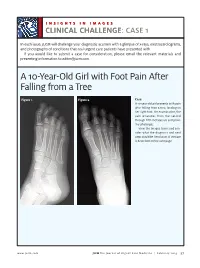
A 10-Year-Old Girl with Foot Pain After Falling from a Tree
INSIGHTS IN IMAGES CLINICAL CHALLENGECHALLENGE: CASE 1 In each issue, JUCM will challenge your diagnostic acumen with a glimpse of x-rays, electrocardiograms, and photographs of conditions that real urgent care patients have presented with. If you would like to submit a case for consideration, please email the relevant materials and presenting information to [email protected]. A 10-Year-Old Girl with Foot Pain After Falling from a Tree Figure 1. Figure 2. Case A 10-year-old girl presents with pain after falling from a tree, landing on her right foot. On examination, the pain emanates from the second through fifth metatarsals and proxi- mal phalanges. View the images taken and con- sider what the diagnosis and next steps would be. Resolution of the case is described on the next page. www.jucm.com JUCM The Journal of Urgent Care Medicine | February 2019 37 INSIGHTS IN IMAGES: CLINICAL CHALLENGE THE RESOLUTION Figure 1. Mediastinal air Figure 1. Differential Diagnosis Pearls for Urgent Care Management and Ⅲ Fracture of the distal fourth metatarsal Considerations for Transfer Ⅲ Plantar plate disruption Ⅲ Emergent transfer should be considered with associated neu- Ⅲ Sesamoiditis rologic deficit, compartment syndrome, open fracture, or vas- Ⅲ Turf toe cular compromise Ⅲ Referral to an orthopedist is warranted in the case of an in- Diagnosis tra-articular fracture, or with Lisfranc ligament injury or ten- Angulation of the distal fourth metatarsal metaphyseal cortex derness over the Lisfranc ligament and hairline lucency consistent with fracture. Acknowledgment: Images courtesy of Teleradiology Associates. Learnings/What to Look for Ⅲ Proximal metatarsal fractures are most often caused by crush- ing or direct blows Ⅲ In athletes, an axial load placed on a plantar-flexed foot should raise suspicion of a Lisfranc injury 38 JUCM The Journal of Urgent Care Medicine | February 2019 www.jucm.com INSIGHTS IN IMAGES CLINICAL CHALLENGE: CASE 2 A 55-Year-Old Man with 3 Hours of Epigastric Pain 55 years PR 249 QRSD 90 QT 471 QTc 425 AXES P 64 QRS -35 T 30 Figure 1. -

Genetic Mechanisms of Disease in Children: a New Look
Genetic Mechanisms of Disease in Children: A New Look Laurie Demmer, MD Tufts Medical Center and the Floating Hospital for Children Boston, MA Traditionally genetic disorders have been linked to the ‘one gene-one protein-one disease’ hypothesis. However recent advances in the field of molecular biology and biotechnology have afforded us the opportunity to greatly expand our knowledge of genetics, and we now know that the mechanisms of inherited disorders are often significantly more complex, and consequently, much more intriguing, than originally thought. Classical mendelian disorders with relatively simple genetic mechanisms do exist, but turn out to be far more rare than originally thought. All patients with sickle cell disease for example, carry the same A-to-T point mutation in the sixth codon of the beta globin gene. This results in a glutamate to valine substitution which changes the shape and the function of the globin molecule in a predictable way. Similarly, all patients with achondroplasia have a single base pair substitution at nucleotide #1138 of the FGFR3 gene. On the other hand, another common inherited disorder, cystic fibrosis, is known to result from changes in a specific transmembrane receptor (CFTR), but over 1000 different disease-causing mutations have been reported in this single gene. Since most commercial labs only test for between 23-100 different mutations, interpreting CFTR mutation testing is significantly complicated by the known risk of false negative results. Many examples of complex, or non-Mendelian, inheritance are now known to exist and include disorders of trinucleotide repeats, errors in imprinting, and gene dosage effects. -

Blueprint Genetics Hereditary Leukemia Panel
Hereditary Leukemia Panel Test code: ON0101 Is a 41 gene panel that includes assessment of non-coding variants. Is ideal for patients with a personal history of a syndrome that confers an increased risk of leukemia or patients with a family history of a syndrome that confers an increased risk of leukemia. About Hereditary Leukemia An inherited predisposition to hematological malignancies, namely acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), and bone marrow myelodysplastic syndrome (MDS) may be associated with syndromic features or occur as the principal clinical feature. MDSs and AMLs can occur in the context of syndromic bone marrow failure (eg. dyskeratosis congenita, Fanconi anemia). Other hereditary syndromes with an increased risk of leukemia include Li-Fraumeni syndrome (TP53), ataxia telangiectasia (ATM), Bloom syndrome (BLM), neurofibromatosis type 1 (NF1) and less frequently Noonan syndrome (PTPN11, CBL). Some reports have also shown an association of biallelic germline mutations in constitutional mismatch repair-deficiency syndrome genes, MLH1, MSH2, MSH6, and PMS2 with the development of ALL. Isolated hematological malignancies are associated with germline mutations in RUNX1 (familial platelet syndrome with predisposition to acute myelogenous leukemia), CEBPA (familial AML), GATA2 (GATA2-associated syndromes) and DDX41(DDX41 -related myeloid neoplasms). There is a rapidly expanding list of germline mutations associated with increased risks for myeloid malignancies and inherited predisposition to hematologic malignancies may be more common than has been thought. Many different genetic defects associated with the development of leukemia have been described but the common underlying mechanism is a dysfunctional DNA damage response. Recognition of an inherited cause provides a specific molecular diagnosis and helps to guide treatment, understand unique disease features, prognosis and other organ systems that may be involved, and identify others in the family who may be at risk. -

Segmental Overvekst Og Vaskulærmalformasjoner V02
2/1/2021 Segmental overvekst og vaskulærmalformasjoner v02 Avdeling for medisinsk genetikk Segmental overvekst og vaskulærmalformasjoner Genpanel, versjon v02 * Enkelte genomiske regioner har lav eller ingen sekvensdekning ved eksomsekvensering. Dette skyldes at de har stor likhet med andre områder i genomet, slik at spesifikk gjenkjennelse av disse områdene og påvisning av varianter i disse områdene, blir vanskelig og upålitelig. Disse genetiske regionene har vi identifisert ved å benytte USCS segmental duplication hvor områder større enn 1 kb og ≥90% likhet med andre regioner i genomet, gjenkjennes (https://genome.ucsc.edu). Vi gjør oppmerksom på at ved identifiseringav ekson oppstrøms for startkodon kan eksonnummereringen endres uten at transkript ID endres. Avdelingens websider har en full oversikt over områder som er affisert av segmentale duplikasjoner. ** Transkriptets kodende ekson. Ekson Gen Gen affisert (HGNC (HGNC Transkript Ekson** Fenotype av symbol) ID) segdup* ACVRL1 175 NM_000020.3 2-10 Telangiectasia, hereditary hemorrhagic, type 2 OMIM ADAMTS3 219 NM_014243.3 1-22 Hennekam lymphangiectasia- lymphedema syndrome 3 OMIM AKT1 391 NM_005163.2 2-14 Cowden syndrome 6 OMIM Proteus syndrome, somatic OMIM AKT2 392 NM_001626.6 2-14 Diabetes mellitus, type II OMIM Hypoinsulinemic hypoglycemia with hemihypertrophy OMIM AKT3 393 NM_005465.7 2-14 Megalencephaly-polymicrogyria- polydactyly-hydrocephalus syndrome 2 OMIM file:///data/SegOv_v02-web.html 1/7 2/1/2021 Segmental overvekst og vaskulærmalformasjoner v02 Ekson Gen Gen affisert (HGNC (HGNC -

Master List of References ACADVL 1
Last Revision: 6/2017 DEPARTMENT OF PATHOLOGY AND LABORATORY MEDICINE PRECISION DIAGNOSTICS – INHERITED DISEASE Master List of References ACADVL 1. Mathur A, Sims HF, Gopalakrishnan D et al. Molecular heterogeneity in very-long-chain acyl-CoA dehydrogenase deficiency causing pediatric cardiomyopathy and sudden death. Circulation 1999;99:1337-43. [PMID: 10077518] 2. Vianey-Saban C, Divry P, Brivet M et al. Mitochondrial very-long-chain acyl-coenzyme A dehydrogenase deficiency: Clinical characteristics and diagnostic considerations in 30 patients. Clin Chim Acta 1998;269:43-62. [PMID: 9498103] AR 1. Giagulli VA, Carbone MD, De Pergola G, et al. Could androgen receptor gene CAG tract polymorphism affect spermatogenesis in men with idiopathic infertility? J Assist Reprod Genet 2014;31(6):689-97. [PMID: 24691874] 2. Gottlieb B, Beitel LK, Nadarajah A, et al. The androgen receptor gene mutations database (ARDB): 2012 update. Hum Mutat. 2012;33:887-94. [PMID: 22334387] Array CGH 1. Boone PM, Bacino CA, Shaw CA et al., Detection of clinically relevant exonic copy-number changes by array CGH. Hum Mutat 2010;31(12):1326-1342. [PMID: 20848651] CFTR 1. Collaco AM, Geibel P, Lee BS et al. Functional vacuolar ATPase (V-ATPase) proton pumps traffic to the enterocyte brush border membrane and require CFTR. Am J Physiol Cell Physiol 2013;305(9):C981-996. [PMID: 23986201] 2. Lu S, Yang X, Li X et al. Different cystic fibrosis transmembrane coductance regulator mutations in Chinese men with congenital bilateral absence of vas deferens and other acquired obtructive azoospermia. Urology 2013;82(4):824-828. [PMID: 23953609] 3. Sosnay PR, Siklosi KR, Van Goor F et al. -

The Atypical Guanine-Nucleotide Exchange Factor, Dock7, Negatively Regulates Schwann Cell Differentiation and Myelination
The Journal of Neuroscience, August 31, 2011 • 31(35):12579–12592 • 12579 Cellular/Molecular The Atypical Guanine-Nucleotide Exchange Factor, Dock7, Negatively Regulates Schwann Cell Differentiation and Myelination Junji Yamauchi,1,3,5 Yuki Miyamoto,1 Hajime Hamasaki,1,3 Atsushi Sanbe,1 Shinji Kusakawa,1 Akane Nakamura,2 Hideki Tsumura,2 Masahiro Maeda,4 Noriko Nemoto,6 Katsumasa Kawahara,5 Tomohiro Torii,1 and Akito Tanoue1 1Department of Pharmacology and 2Laboratory Animal Resource Facility, National Research Institute for Child Health and Development, Setagaya, Tokyo 157-8535, Japan, 3Department of Biological Sciences, Tokyo Institute of Technology, Midori, Yokohama 226-8501, Japan, 4IBL, Ltd., Fujioka, Gumma 375-0005, Japan, and 5Department of Physiology and 6Bioimaging Research Center, Kitasato University School of Medicine, Sagamihara, Kanagawa 252-0374, Japan In development of the peripheral nervous system, Schwann cells proliferate, migrate, and ultimately differentiate to form myelin sheath. In all of the myelination stages, Schwann cells continuously undergo morphological changes; however, little is known about their underlying molecular mechanisms. We previously cloned the dock7 gene encoding the atypical Rho family guanine-nucleotide exchange factor (GEF) and reported the positive role of Dock7, the target Rho GTPases Rac/Cdc42, and the downstream c-Jun N-terminal kinase in Schwann cell migration (Yamauchi et al., 2008). We investigated the role of Dock7 in Schwann cell differentiation and myelination. Knockdown of Dock7 by the specific small interfering (si)RNA in primary Schwann cells promotes dibutyryl cAMP-induced morpholog- ical differentiation, indicating the negative role of Dock7 in Schwann cell differentiation. It also results in a shorter duration of activation of Rac/Cdc42 and JNK, which is the negative regulator of myelination, and the earlier activation of Rho and Rho-kinase, which is the positive regulator of myelination. -

Exostoses, Enchondromatosis and Metachondromatosis; Diagnosis and Management
Acta Orthop. Belg., 2016, 82, 102-105 ORIGINAL STUDY Exostoses, enchondromatosis and metachondromatosis; diagnosis and management John MCFARLANE, Tim KNIGHT, Anubha SINHA, Trevor COLE, Nigel KIELY, Rob FREEMAN From the Department of Orthopaedics, Robert Jones Agnes Hunt Hospital, Oswestry, UK We describe a 5 years old girl who presented to the region of long bones and are composed of a carti- multidisciplinary skeletal dysplasia clinic following lage lump outside the bone which may be peduncu- excision of two bony lumps from her fingers. Based on lated or sessile, the knee is the most common clinical examination, radiolographs and histological site (1,10). An isolated exostosis is a common inci- results an initial diagnosis of hereditary multiple dental finding rarely requiring treatment. Disorders exostosis (HME) was made. Four years later she developed further lumps which had the radiological associated with exostoses include HME, Langer- appearance of enchondromas. The appearance of Giedion syndrome, Gardner syndrome and meta- both exostoses and enchondromas suggested a possi- chondromatosis. ble diagnosis of metachondromatosis. Genetic testing Enchondroma are the second most common be- revealed a splice site mutation at the end of exon 11 on nign bone tumour characterised by the formation of the PTPN11 gene, confirming the diagnosis of meta- hyaline cartilage in the medulla of a bone. It occurs chondromatosis. While both single or multiple exosto- most frequently in the hand (60%) and then the feet. ses and enchondromas occur relatively commonly on The typical radiological features are of a well- their own, the appearance of multiple exostoses and defined lucent defect with endosteal scalloping and enchondromas together is rare and should raise the differential diagnosis of metachondromatosis. -

Phenotypic and Genotypic Characterisation of Noonan-Like
1of5 ELECTRONIC LETTER J Med Genet: first published as 10.1136/jmg.2004.024091 on 2 February 2005. Downloaded from Phenotypic and genotypic characterisation of Noonan-like/ multiple giant cell lesion syndrome J S Lee, M Tartaglia, B D Gelb, K Fridrich, S Sachs, C A Stratakis, M Muenke, P G Robey, M T Collins, A Slavotinek ............................................................................................................................... J Med Genet 2005;42:e11 (http://www.jmedgenet.com/cgi/content/full/42/2/e11). doi: 10.1136/jmg.2004.024091 oonan-like/multiple giant cell lesion syndrome (NL/ MGCLS; OMIM 163955) is a rare condition1–3 with Key points Nphenotypic overlap with Noonan’s syndrome (OMIM 163950) and cherubism (OMIM 118400) (table 1). N Noonan-like/multiple giant cell lesion syndrome (NL/ Recently, missense mutations in the PTPN11 gene on MGCLS) has clinical similarities with Noonan’s syn- chromosome 12q24.1 have been identified as the cause of drome and cherubism. It is unclear whether it is a Noonan’s syndrome in 45% of familial and sporadic cases,45 distinct entity or a variant of Noonan’s syndrome or indicating genetic heterogeneity within the syndrome. In the cherubism. 5 study by Tartaglia et al, there was a family in which three N Three unrelated patients with NL/MGCLS were char- members had features of Noonan’s syndrome; two of these acterised, two of whom were found to have mutations had incidental mandibular giant cell lesions.3 All three in the PTPN11 gene, the mutation found in 45% of members were found to have a PTPN11 mutation known to patients with Noonan’s syndrome. -

Megalencephaly and Macrocephaly
277 Megalencephaly and Macrocephaly KellenD.Winden,MD,PhD1 Christopher J. Yuskaitis, MD, PhD1 Annapurna Poduri, MD, MPH2 1 Department of Neurology, Boston Children’s Hospital, Boston, Address for correspondence Annapurna Poduri, Epilepsy Genetics Massachusetts Program, Division of Epilepsy and Clinical Electrophysiology, 2 Epilepsy Genetics Program, Division of Epilepsy and Clinical Department of Neurology, Fegan 9, Boston Children’s Hospital, 300 Electrophysiology, Department of Neurology, Boston Children’s Longwood Avenue, Boston, MA 02115 Hospital, Boston, Massachusetts (e-mail: [email protected]). Semin Neurol 2015;35:277–287. Abstract Megalencephaly is a developmental disorder characterized by brain overgrowth secondary to increased size and/or numbers of neurons and glia. These disorders can be divided into metabolic and developmental categories based on their molecular etiologies. Metabolic megalencephalies are mostly caused by genetic defects in cellular metabolism, whereas developmental megalencephalies have recently been shown to be caused by alterations in signaling pathways that regulate neuronal replication, growth, and migration. These disorders often lead to epilepsy, developmental disabilities, and Keywords behavioral problems; specific disorders have associations with overgrowth or abnor- ► megalencephaly malities in other tissues. The molecular underpinnings of many of these disorders are ► hemimegalencephaly now understood, providing insight into how dysregulation of critical pathways leads to ► -

Integrative Genomics Analysis of Eqtl and GWAS Summary Data Identifies
Bioscience Reports (2020) 40 BSR20193185 https://doi.org/10.1042/BSR20193185 Research Article Integrative genomics analysis of eQTL and GWAS summary data identifies PPP1CB as a novel bone mineral density risk genes Yu Zhai1,2,*,LuYu1,*, Yang Shao2 and Jianwei Wang2 Downloaded from http://portlandpress.com/bioscirep/article-pdf/40/4/BSR20193185/872280/bsr-2019-3185.pdf by guest on 01 October 2021 1Changzhou Hospital Affiliated to Nanjing University of Chinese Medicine, Changzhou 213003, Jiangsu, China; 2Wuxi Hospital Affiliated to Nanjing University of Chinese Medicine, Wuxi 214071, Jiangsu, China Correspondence: Jianwei Wang ([email protected]) In recent years, multiple genome-wide association studies (GWAS) have identified numerous susceptibility variants and risk genes that demonstrate significant associations with bone mineral density (BMD). However, exploring how these genetic variants contribute risk to BMD remains a major challenge. We systematically integrated two independent expression quantitative trait loci (eQTL) data (N = 1890) and GWAS summary statistical data of BMD (N = 142,487) using Sherlock integrative analysis to reveal whether expression-associated vari- ants confer risk to BMD. By using Sherlock integrative analysis and MAGMA gene-based analysis, we found there existed 36 promising genes, for example, PPP1CB, XBP1, and FDFT1, whose expression alterations may contribute susceptibility to BMD. Through a protein–protein interaction (PPI) network analysis, we further prioritized the PPP1CB as a hub gene that has interactions with predicted genes and BMD-associated genes. Two eS- −17 −11 NPs of rs9309664 (PeQTL = 1.42 × 10 and PGWAS = 1.40 × 10 ) and rs7475 (PeQTL = −6 −7 2.10 × 10 and PGWAS = 1.70 × 10 )inPPP1CB were identified to be significantly as- sociated with BMD risk. -

Cardiomyopathy Precision Panel Overview Indications
Cardiomyopathy Precision Panel Overview Cardiomyopathies are a group of conditions with a strong genetic background that structurally hinder the heart to pump out blood to the rest of the body due to weakness in the heart muscles. These diseases affect individuals of all ages and can lead to heart failure and sudden cardiac death. If there is a family history of cardiomyopathy it is strongly recommended to undergo genetic testing to be aware of the family risk, personal risk, and treatment options. Most types of cardiomyopathies are inherited in a dominant manner, which means that one altered copy of the gene is enough for the disease to present in an individual. The symptoms of cardiomyopathy are variable, and these diseases can present in different ways. There are 5 types of cardiomyopathies, the most common being hypertrophic cardiomyopathy: 1. Hypertrophic cardiomyopathy (HCM) 2. Dilated cardiomyopathy (DCM) 3. Restrictive cardiomyopathy (RCM) 4. Arrhythmogenic Right Ventricular Cardiomyopathy (ARVC) 5. Isolated Left Ventricular Non-Compaction Cardiomyopathy (LVNC). The Igenomix Cardiomyopathy Precision Panel serves as a diagnostic and tool ultimately leading to a better management and prognosis of the disease. It provides a comprehensive analysis of the genes involved in this disease using next-generation sequencing (NGS) to fully understand the spectrum of relevant genes. Indications The Igenomix Cardiomyopathy Precision Panel is indicated in those cases where there is a clinical suspicion of cardiomyopathy with or without the following manifestations: - Shortness of breath - Fatigue - Arrythmia (abnormal heart rhythm) - Family history of arrhythmia - Abnormal scans - Ventricular tachycardia - Ventricular fibrillation - Chest Pain - Dizziness - Sudden cardiac death in the family 1 Clinical Utility The clinical utility of this panel is: - The genetic and molecular diagnosis for an accurate clinical diagnosis of a patient with personal or family history of cardiomyopathy, channelopathy or sudden cardiac death. -

Hepatitis C Virus As a Unique Human Model Disease to Define
viruses Review Hepatitis C Virus as a Unique Human Model Disease to Define Differences in the Transcriptional Landscape of T Cells in Acute versus Chronic Infection David Wolski and Georg M. Lauer * Liver Center at the Gastrointestinal Unit, Department of Medicine, Massachusetts General Hospital and Harvard Medical School, Boston, MA 02114, USA * Correspondence: [email protected]; Tel.: +1-617-724-7515 Received: 27 June 2019; Accepted: 23 July 2019; Published: 26 July 2019 Abstract: The hepatitis C virus is unique among chronic viral infections in that an acute outcome with complete viral elimination is observed in a minority of infected patients. This unique feature allows direct comparison of successful immune responses with those that fail in the setting of the same human infection. Here we review how this scenario can be used to achieve better understanding of transcriptional regulation of T-cell differentiation. Specifically, we discuss results from a study comparing transcriptional profiles of hepatitis C virus (HCV)-specific CD8 T-cells during early HCV infection between patients that do and do not control and eliminate HCV. Identification of early gene expression differences in key T-cell differentiation molecules as well as clearly distinct transcriptional networks related to cell metabolism and nucleosomal regulation reveal novel insights into the development of exhausted and memory T-cells. With additional transcriptional studies of HCV-specific CD4 and CD8 T-cells in different stages of infection currently underway, we expect HCV infection to become a valuable model disease to study human immunity to viruses. Keywords: viral hepatitis; hepatitis C virus; T cells; transcriptional regulation; transcription factors; metabolism; nucleosome 1.