KEGGID Term Gene 4612 Antigen Processing and HLA-DRB1, HLA
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplementary Materials
DEPs in osteosarcoma cells comparing to osteoblastic cells Biological Process Protein Percentage of Hits metabolic process (GO:0008152) 29.3 29.3% cellular process (GO:0009987) 20.2 20.2% localization (GO:0051179) 9.4 9.4% biological regulation (GO:0065007) 8 8.0% developmental process (GO:0032502) 7.8 7.8% response to stimulus (GO:0050896) 5.6 5.6% cellular component organization (GO:0071840) 5.6 5.6% multicellular organismal process (GO:0032501) 4.4 4.4% immune system process (GO:0002376) 4.2 4.2% biological adhesion (GO:0022610) 2.7 2.7% apoptotic process (GO:0006915) 1.6 1.6% reproduction (GO:0000003) 0.8 0.8% locomotion (GO:0040011) 0.4 0.4% cell killing (GO:0001906) 0.1 0.1% 100.1% Genes 2179Hits 3870 biological adhesion apoptotic process … reproduction (GO:0000003) , 0.8% (GO:0022610) , 2.7% locomotion (GO:0040011) ,… immune system process cell killing (GO:0001906) , 0.1% (GO:0002376) , 4.2% multicellular organismal process (GO:0032501) , metabolic process 4.4% (GO:0008152) , 29.3% cellular component organization (GO:0071840) , 5.6% response to stimulus (GO:0050896), 5.6% developmental process (GO:0032502) , 7.8% biological regulation (GO:0065007) , 8.0% cellular process (GO:0009987) , 20.2% localization (GO:0051179) , 9. -

Supplementary Information Material and Methods
MCT-11-0474 BKM120: a potent and specific pan-PI3K inhibitor Supplementary Information Material and methods Chemicals The EGFR inhibitor NVP-AEE788 (Novartis), the Jak inhibitor I (Merck Calbiochem, #420099) and anisomycin (Alomone labs, # A-520) were prepared as 50 mM stock solutions in 100% DMSO. Doxorubicin (Adriablastin, Pfizer), EGF (Sigma Ref: E9644), PDGF (Sigma, Ref: P4306) and IL-4 (Sigma, Ref: I-4269) stock solutions were prepared as recommended by the manufacturer. For in vivo administration: Temodal (20 mg Temozolomide capsules, Essex Chemie AG, Luzern) was dissolved in 4 mL KZI/glucose (20/80, vol/vol); Taxotere was bought as 40 mg/mL solution (Sanofi Aventis, France), and prepared in KZI/glucose. Antibodies The primary antibodies used were as follows: anti-S473P-Akt (#9271), anti-T308P-Akt (#9276,), anti-S9P-GSK3β (#9336), anti-T389P-p70S6K (#9205), anti-YP/TP-Erk1/2 (#9101), anti-YP/TP-p38 (#9215), anti-YP/TP-JNK1/2 (#9101), anti-Y751P-PDGFR (#3161), anti- p21Cip1/Waf1 (#2946), anti-p27Kip1 (#2552) and anti-Ser15-p53 (#9284) antibodies were from Cell Signaling Technologies; anti-Akt (#05-591), anti-T32P-FKHRL1 (#06-952) and anti- PDGFR (#06-495) antibodies were from Upstate; anti-IGF-1R (#SC-713) and anti-EGFR (#SC-03) antibodies were from Santa Cruz; anti-GSK3α/β (#44610), anti-Y641P-Stat6 (#611566), anti-S1981P-ATM (#200-301), anti-T2609 DNA-PKcs (#GTX24194) and anti- 1 MCT-11-0474 BKM120: a potent and specific pan-PI3K inhibitor Y1316P-IGF-1R were from Bio-Source International, Becton-Dickinson, Rockland, GenTex and internal production, respectively. The 4G10 antibody was from Millipore (#05-321MG). -
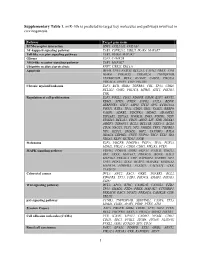
1 Supplementary Table 1. Mir-10B Is Predicted to Target Key Molecules
Supplementary Table 1. miR-10b is predicted to target key molecules and pathways involved in carcinogenesis. Pathway Target gene name ECM-receptor interaction SDC1, COL24A1, COL4A4 NF-kappa B signaling pathway TAB1, CSNK2A2, UBE2I, IRAK4, MAP3K7 Toll-like receptor signaling pathway TAB1, IRAK4, MAP3K7 Glioma E2F3, CAMK2B NOD-like receptor signaling pathway TAB1, MAP3K7 Ubiquitin mediated proteolysis RNF7, UBE2I, ERCC8 Apoptosis DFFB, TP53, FASLG, BCL2L1, CAPN2, PRKX, ATM, IRAK4, PRKACG, PRKAR2A, TNFRSF10B, TNFRSF10D, BCL2, IL1RAP, CASP8, PIK3CA, PRKACA, APAF1, CHP, PIK3R3 Chronic myeloid leukemia E2F3, BCR, GRB2, TGFBR1, CBL, TP53, CDK6, BCL2L1, GAB2, PIK3CA, MDM2, SHC1, PIK3R3, CRK Regulation of cell proliferation E2F3, FOSL2, CDX2, PDGFB, OSMR, E2F7, ARNT2, RBM5, STRN, PTEN, S1PR2, CUL3, BDNF, SERPINE1, SHC1, ASPH, ITCH, SPN, CCDC88A, FOXJ1, RXRA, TP53, CDK6, IRS1, VASH2, RBBP9, VASH1, ADRB2, PDGFRA, MDM2, ADAMTS1, EIF2AK2, EIF5A2, ICOSLG, ING5, FGFR3, NDN, ST8SIA1, BCL2L1, CDH5, ARNT, LIF, VDR, HOXA3, AGGF1, TSPAN31, BCL2, BCL11B, NKX3-1, BCL6, CD28, NACC1, FLT1, NF2, JARID2, TBX5, TGFBR1, NF1, KLF11, SMAD2, IGF2, TAX1BP3, BTLA, HDAC4, LEPRE1, CNTF, NUP62, TSC1, ETS1, ID4, NR5A2, KLF4, KCTD11, NFIB Melanoma E2F3, PDGFB, PDGFRA, FGF11, TP53, FGF23, MDM2, PIK3CA, CDK6, CDH1, PIK3R3, PTEN MAPK signaling pathway FGFR3, PDGFB, GRB2, FGF11, FASLG, GNG12, SRF, PRKX, MAP3K7, PRKACG, BDNF, RAC3, MAP3K2, PRKACA, CHP, RAPGEF2, TGFBR1, NF1, TP53, FGF23, STK4, DUSP5, MAP4K4, RPS6KA2, MAPK14, PDGFRA, PLA2G3, CACNA1C, CRK, PLA2G2F Colorectal cancer -

Swine in Biomedical Research Conference 2005
Swine in Biomedical Research Conference 2005 www.swinegenomics.com Organizing Committee: Lawrence B. Schook, Ph.D. (Chair) Dept. Animal Sciences, Veterinary Pathobiology & Nutritional Sciences, Institute for Genomic Biology, University of Illinois Christopher Counter, Ph.D. Dept. of Oncology and Cancer Biology, Duke University Medical Center Eric Forsberg, Ph.D Vice President of Business Development, Infigen Merete Fredholm, D.V.M., Ph.D., Dr. Vet. Sci., Royal Veterinary and Agricultural Univ.Inst. Animal Science & Animal Health Medicine Thalachallour Mohanakumar, Ph.D. Dept. of Surgery, Pathology and Immunology, Washington University School of Medicine Randall Prather, Ph.D. Department of Reproductive Biotechnology, University of Missouri Steven Niemi, D.V.M. Center for Comparative Medicine, Massachusetts General Hospital Hiroshi Yasue, Ph.D. National Institute of Agrobiological Sciences, Sukuba, Japan University of Illinois Hosting Committee: Jonathan Beever, Animal Sciences Sharon Donovan, Food Science & Human Nutrition H. Rex Gaskins, Animal Sciences Russell Jamison, Materials Science & Engineering Lawrence Schook, Animal Sciences Kelley Tappenden, Food Science & Human Nutrition Michael Tumbleson, Agricultural Engineering (honorary chair) Matthew Wheeler, Animal Sciences Federico Zuckermann, Veterinary Pathobiology Poster Session Moderators: Bioengineering, Russ Jamison, UIUC Immunology & Infectious Diseases, Federico Zuckermann, UIUC Transplantation (allo & xeno), Doug Smith, Baylor & Mark Rutherford, U.Minn. Nutrition (Obesity and Diabetes), Sharon Donovan, UIUC Genomics and Cloning, Jon Beever, UIUC & Max Rothschild, ISU Cardiovascular, Rex Gaskins, UIUC Physiology, Jack Odle, NCSU Cancer, Craig Beattie, UNR Clinical Models, Steve Niemi, Harvard Sponsored by: Institute for Genomic Biology, College of Agricultural, Consumer, and Environmental Services, College of Liberal Arts & Sciences, College of Veterinary Medicine, and Office of the Vice-Chancellor of Research at the University of Illinois at Urbana-Champaign National Institutes for Health (Grant no. -

ADVISORY COMMISSION on CHILDHOOD VACCINES TABLE of CONTENTS December 8, 2017
ADVISORY COMMISSION ON CHILDHOOD VACCINES TABLE OF CONTENTS December 8, 2017 TAB • ACCV Agenda 1 • ACCV Charter • ACCV Roster • 2017 Meeting Dates • Meeting Minutes 2 o Draft Minutes – September 8, 2017 • Vaccine Injury Compensation Trust Fund Statement 3 o Vaccine Injury Compensation Trust Fund Summary Sheet for the Period of 10/1/2016 – 9/30/2017 • VICP Data and Statistics 4 • Meeting Presentations & Updates 5 o Report from the Division of Injury Compensation Programs 5.1 o Report from the Department of Justice 5.2 o Petitions to Add Injuries to the Vaccine Injury Table Introduction 5.3 o Petition to Add Tics as an Injury to the Vaccine Injury Table 5.4 o Petition to Add Asthma as an Injury to the Vaccine Injury Table 5.5 5.6 o Petition to Add Pediatric Autoimmune Neuropsychiatric Syndrome (PANS), Pediatric Infection-Triggered Autoimmune Neuropsychiatric Disorders (PITANDS), and Pediatric Autoimmune Neuropsychiatric Disorders (PANDAS) as Injuries to the Vaccine Injury Table o Petition to Add Experimental Autoimmune Encephalomyelitis (EAE) and/or 5.7 Acute Demyelinating Encephalomyelitis (ADEM) as injuries to the Vaccine Injury Table 5.8 o Update on the Immunization Safety Office Vaccine Activities (CDC) o Update on the National Institute of Allergy and Infectious Diseases Vaccine 5.9 Activities (NIH) o Update on the Center for Biologics, Evaluation and Research Vaccine 5.10 Activities (FDA) 5.11 o Update from the National Vaccine Program Office • Program Related Articles 6 6.1 o Popular Science, “Why Are We So Bad At Producing The Right -

The P90 RSK Family Members: Common Functions and Isoform Specificity
Published OnlineFirst August 22, 2013; DOI: 10.1158/0008-5472.CAN-12-4448 Cancer Review Research The p90 RSK Family Members: Common Functions and Isoform Specificity Romain Lara, Michael J. Seckl, and Olivier E. Pardo Abstract The p90 ribosomal S6 kinases (RSK) are implicated in various cellular processes, including cell proliferation, survival, migration, and invasion. In cancer, RSKs modulate cell transformation, tumorigenesis, and metastasis. Indeed, changes in the expression of RSK isoforms have been reported in several malignancies, including breast, prostate, and lung cancers. Four RSK isoforms have been identified in humans on the basis of their high degree of sequence homology. Although this similarity suggests some functional redundancy between these proteins, an increasing body of evidence supports the existence of isoform-based specificity among RSKs in mediating particular cellular processes. This review briefly presents the similarities between RSK family members before focusing on the specific function of each of the isoforms and their involvement in cancer progression. Cancer Res; 73(17); 1–8. Ó2013 AACR. Introduction subsequently cloned throughout the Metazoan kingdom (2). The extracellular signal–regulated kinase (ERK)1/2 pathway The genomic analysis of several cancer types suggests that fi is involved in key cellular processes, including cell prolifera- these genes are not frequently ampli ed or mutated, with some tion, differentiation, survival, metabolism, and migration. notable exceptions (e.g., in the case of hepatocellular carcino- More than 30% of all human cancers harbor mutations within ma; ref. 6). Table 1 summarizes reported genetic changes in this pathway, mostly resulting in gain of function and conse- RSK genes. -

Regulates Target Gene Transcription Via Single-Stranded DNA Response Elements
TLS/FUS (translocated in liposarcoma/fused in sarcoma) regulates target gene transcription via single-stranded DNA response elements Adelene Y. Tan1, Todd R. Riley, Tristan Coady, Harmen J. Bussemaker, and James L. Manley2 Department of Biological Sciences, Columbia University, New York, NY 10027 Contributed by James L. Manley, February 29, 2012 (sent for review December 9, 2011) TLS/FUS (TLS) is a multifunctional protein implicated in a wide range TLS has also been linked to splicing. It contains an RNP-type of cellular processes, including transcription and mRNA processing, RNA-binding domain and associates directly with SR protein as well as in both cancer and neurological disease. However, little is splicing factors (11). TET proteins have been detected in spliceo- currently known about TLS target genes and how they are recog- somes (12), and TLS was found associated with RNAP II and nized. Here, we used ChIP and promoter microarrays to identify snRNPs in a transcription and splicing complex in vitro (13). It is genes potentially regulated by TLS. Among these genes, we detected unclear whether and how TLS recruits splicing factors to sites of a number that correlate with previously known functions of TLS, active transcription, but one possibility is through its interaction and confirmed TLS occupancy at several of them by ChIP. We also with TBP and the TFIID complex. detected changes in mRNA levels of these target genes in cells where Here we provide insight into TLS regulation of RNAP II-tran- scribed genes. We used ChIP followed by promoter microarray TLS levels were altered, indicative of both activation and repression. -

CPTC-MAPK3-1 (CAB079934) Immunohistochemistry
CPTC-MAPK3-1 (CAB079934) Uniprot ID: P27361 Protein name: MK03_HUMAN Full name: Mitogen-activated protein kinase 3 Function: Serine/threonine kinase which acts as an essential component of the MAP kinase signal transduction pathway. MAPK1/ERK2 and MAPK3/ERK1 are the 2 MAPKs which play an important role in the MAPK/ERK cascade. They participate also in a signaling cascade initiated by activated KIT and KITLG/SCF. Depending on the cellular context, the MAPK/ERK cascade mediates diverse biological functions such as cell growth, adhesion, survival and differentiation through the regulation of transcription, translation, cytoskeletal rearrangements. The MAPK/ERK cascade plays also a role in initiation and regulation of meiosis, mitosis, and postmitotic functions in differentiated cells by phosphorylating a number of transcription factors. About 160 substrates have already been discovered for ERKs. Many of these substrates are localized in the nucleus, and seem to participate in the regulation of transcription upon stimulation. However, other substrates are found in the cytosol as well as in other cellular organelles, and those are responsible for processes such as translation, mitosis and apoptosis. Moreover, the MAPK/ERK cascade is also involved in the regulation of the endosomal dynamics, including lysosome processing and endosome cycling through the perinuclear recycling compartment (PNRC); as well as in the fragmentation of the Golgi apparatus during mitosis. The substrates include transcription factors (such as ATF2, BCL6, ELK1, ERF, FOS, HSF4 or SPZ1), cytoskeletal elements (such as CANX, CTTN, GJA1, MAP2, MAPT, PXN, SORBS3 or STMN1), regulators of apoptosis (such as BAD, BTG2, CASP9, DAPK1, IER3, MCL1 or PPARG), regulators of translation (such as EIF4EBP1) and a variety of other signaling-related molecules (like ARHGEF2, FRS2 or GRB10). -

Functional Analysis of Structural Variation in the 2D and 3D Human Genome
FUNCTIONAL ANALYSIS OF STRUCTURAL VARIATION IN THE 2D AND 3D HUMAN GENOME by Conor Mitchell Liam Nodzak A dissertation submitted to the faculty of The University of North Carolina at Charlotte in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Bioinformatics and Computational Biology Charlotte 2019 Approved by: Dr. Xinghua Mindy Shi Dr. Rebekah Rogers Dr. Jun-tao Guo Dr. Adam Reitzel ii c 2019 Conor Mitchell Liam Nodzak ALL RIGHTS RESERVED iii ABSTRACT CONOR MITCHELL LIAM NODZAK. Functional analysis of structural variation in the 2D and 3D human genome. (Under the direction of DR. XINGHUA MINDY SHI) The human genome consists of over 3 billion nucleotides that have an average distance of 3.4 Angstroms between each base, which equates to over two meters of DNA contained within the 125 µm3 volume diploid cell nuclei. The dense compaction of chromatin by the supercoiling of DNA forms distinct architectural modules called topologically associated domains (TADs), which keep protein-coding genes, noncoding RNAs and epigenetic regulatory elements in close nuclear space. It has recently been shown that these conserved chromatin structures may contribute to tissue-specific gene expression through the encapsulation of genes and cis-regulatory elements, and mutations that affect TADs can lead to developmental disorders and some forms of cancer. At the population-level, genomic structural variation contributes more to cumulative genetic difference than any other class of mutation, yet much remains to be studied as to how structural variation affects TADs. Here, we study the func- tional effects of structural variants (SVs) through the analysis of chromatin topology and gene activity for three trio families sampled from genetically diverse popula- tions from the Human Genome Structural Variation Consortium. -

Open Data for Differential Network Analysis in Glioma
International Journal of Molecular Sciences Article Open Data for Differential Network Analysis in Glioma , Claire Jean-Quartier * y , Fleur Jeanquartier y and Andreas Holzinger Holzinger Group HCI-KDD, Institute for Medical Informatics, Statistics and Documentation, Medical University Graz, Auenbruggerplatz 2/V, 8036 Graz, Austria; [email protected] (F.J.); [email protected] (A.H.) * Correspondence: [email protected] These authors contributed equally to this work. y Received: 27 October 2019; Accepted: 3 January 2020; Published: 15 January 2020 Abstract: The complexity of cancer diseases demands bioinformatic techniques and translational research based on big data and personalized medicine. Open data enables researchers to accelerate cancer studies, save resources and foster collaboration. Several tools and programming approaches are available for analyzing data, including annotation, clustering, comparison and extrapolation, merging, enrichment, functional association and statistics. We exploit openly available data via cancer gene expression analysis, we apply refinement as well as enrichment analysis via gene ontology and conclude with graph-based visualization of involved protein interaction networks as a basis for signaling. The different databases allowed for the construction of huge networks or specified ones consisting of high-confidence interactions only. Several genes associated to glioma were isolated via a network analysis from top hub nodes as well as from an outlier analysis. The latter approach highlights a mitogen-activated protein kinase next to a member of histondeacetylases and a protein phosphatase as genes uncommonly associated with glioma. Cluster analysis from top hub nodes lists several identified glioma-associated gene products to function within protein complexes, including epidermal growth factors as well as cell cycle proteins or RAS proto-oncogenes. -

ERBB2 Influences the Subcellular Localization of the Estrogen
Endocrine-Related Cancer (2008) 15 985–1002 ERBB2 influences the subcellular localization of the estrogen receptor in tamoxifen-resistant MCF-7 cells leading to the activation of AKT and RPS6KA2 Sunil Pancholi, Anne E Lykkesfeldt1, Caroline Hilmi, Susana Banerjee, Alexandra Leary, Suzanne Drury, Stephen Johnston 2,MitchDowsett and Lesley-Ann Martin Institute of Cancer Research, The Breakthrough Breast Cancer Research Centre, 237 Fulham Road, London SW3 6JB, UK 1Institute of Cancer Biology, Danish Cancer Society, Copenhagen, Denmark 2Department of Medicine, Royal Marsden Hospital, Fulham Road, London, UK (Correspondence should be addressed to L-A Martin; Email: [email protected]) Abstract Acquired resistance to endocrine therapies remains a major clinical obstacle in hormone-sensitive breast tumors. We used an MCF-7 breast tumor cell line (TamR-1) resistant to tamoxifen to investigate this mechanism. We demonstrate that TamR-1 express elevated levels of phosphorylated AKT and MAPK3/1-activated RPS6KA2 compared with the parental MCF-7 cell line (MCF-7). There was no change in the level of total ESR between the two cell lines; however, the TamR-1 cells had increased phosphorylation of ESR1 ser167. SiRNA blockade of AKT or MAPK3/1 had little effect on ESR1 ser167 phosphorylation, but a combination of the two siRNAs abrogated this. Co-localization studies revealed an association between ERBB2 and ESR1 in the TamR-1 but not MCF-7 cells. ESR1 was redistributed to extranuclear sites in TamR-1 and was less transcriptionally competent compared with MCF-7 suggesting that nuclear ESR1 activity was suppressed in TamR-1. Tamoxifen resistance in the TamR-1 cells could be partially overcome by the ERBB2 inhibitor AG825 in combination with tamoxifen, and this was associated with re-localization of ESR1 to the nucleus. -

Genome-Wide DNA Methylation Dynamics During Epigenetic
Gómez‑Redondo et al. Clin Epigenet (2021) 13:27 https://doi.org/10.1186/s13148‑021‑01003‑x RESEARCH Open Access Genome‑wide DNA methylation dynamics during epigenetic reprogramming in the porcine germline Isabel Gómez‑Redondo1*† , Benjamín Planells1†, Sebastián Cánovas2,3, Elena Ivanova4, Gavin Kelsey4,5 and Alfonso Gutiérrez‑Adán1 Abstract Background: Prior work in mice has shown that some retrotransposed elements remain substantially methylated during DNA methylation reprogramming of germ cells. In the pig, however, information about this process is scarce. The present study was designed to examine the methylation profles of porcine germ cells during the time course of epigenetic reprogramming. Results: Sows were artifcially inseminated, and their fetuses were collected 28, 32, 36, 39, and 42 days later. At each time point, genital ridges were dissected from the mesonephros and germ cells were isolated through magnetic‑ activated cell sorting using an anti‑SSEA‑1 antibody, and recovered germ cells were subjected to whole‑genome bisulphite sequencing. Methylation levels were quantifed using SeqMonk software by performing an unbiased analysis, and persistently methylated regions (PMRs) in each sex were determined to extract those regions showing 50% or more methylation. Most genomic elements underwent a dramatic loss of methylation from day 28 to day 36, when the lowest levels were shown. By day 42, there was evidence for the initiation of genomic re‑methylation. We identifed a total of 1456 and 1122 PMRs in male and female germ cells, respectively, and large numbers of transpos‑ able elements (SINEs, LINEs, and LTRs) were found to be located within these PMRs. Twenty‑one percent of the introns located in these PMRs were found to be the frst introns of a gene, suggesting their regulatory role in the expression of these genes.