Cell Lines Or Lymphocytes Collected from Blood Via Trizol HDAC4 in Each of These Cases Revealed De Novo Mutations
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

MECHANISMS in ENDOCRINOLOGY: Novel Genetic Causes of Short Stature
J M Wit and others Genetics of short stature 174:4 R145–R173 Review MECHANISMS IN ENDOCRINOLOGY Novel genetic causes of short stature 1 1 2 2 Jan M Wit , Wilma Oostdijk , Monique Losekoot , Hermine A van Duyvenvoorde , Correspondence Claudia A L Ruivenkamp2 and Sarina G Kant2 should be addressed to J M Wit Departments of 1Paediatrics and 2Clinical Genetics, Leiden University Medical Center, PO Box 9600, 2300 RC Leiden, Email The Netherlands [email protected] Abstract The fast technological development, particularly single nucleotide polymorphism array, array-comparative genomic hybridization, and whole exome sequencing, has led to the discovery of many novel genetic causes of growth failure. In this review we discuss a selection of these, according to a diagnostic classification centred on the epiphyseal growth plate. We successively discuss disorders in hormone signalling, paracrine factors, matrix molecules, intracellular pathways, and fundamental cellular processes, followed by chromosomal aberrations including copy number variants (CNVs) and imprinting disorders associated with short stature. Many novel causes of GH deficiency (GHD) as part of combined pituitary hormone deficiency have been uncovered. The most frequent genetic causes of isolated GHD are GH1 and GHRHR defects, but several novel causes have recently been found, such as GHSR, RNPC3, and IFT172 mutations. Besides well-defined causes of GH insensitivity (GHR, STAT5B, IGFALS, IGF1 defects), disorders of NFkB signalling, STAT3 and IGF2 have recently been discovered. Heterozygous IGF1R defects are a relatively frequent cause of prenatal and postnatal growth retardation. TRHA mutations cause a syndromic form of short stature with elevated T3/T4 ratio. Disorders of signalling of various paracrine factors (FGFs, BMPs, WNTs, PTHrP/IHH, and CNP/NPR2) or genetic defects affecting cartilage extracellular matrix usually cause disproportionate short stature. -

Prenatal Ultrasonography of Craniofacial Abnormalities
Prenatal ultrasonography of craniofacial abnormalities Annisa Shui Lam Mak, Kwok Yin Leung Department of Obstetrics and Gynaecology, Queen Elizabeth Hospital, Hong Kong SAR, China REVIEW ARTICLE https://doi.org/10.14366/usg.18031 pISSN: 2288-5919 • eISSN: 2288-5943 Ultrasonography 2019;38:13-24 Craniofacial abnormalities are common. It is important to examine the fetal face and skull during prenatal ultrasound examinations because abnormalities of these structures may indicate the presence of other, more subtle anomalies, syndromes, chromosomal abnormalities, or even rarer conditions, such as infections or metabolic disorders. The prenatal diagnosis of craniofacial abnormalities remains difficult, especially in the first trimester. A systematic approach to the fetal Received: May 29, 2018 skull and face can increase the detection rate. When an abnormality is found, it is important Revised: June 30, 2018 to perform a detailed scan to determine its severity and search for additional abnormalities. Accepted: July 3, 2018 Correspondence to: The use of 3-/4-dimensional ultrasound may be useful in the assessment of cleft palate and Kwok Yin Leung, MBBS, MD, FRCOG, craniosynostosis. Fetal magnetic resonance imaging can facilitate the evaluation of the palate, Cert HKCOG (MFM), Department of micrognathia, cranial sutures, brain, and other fetal structures. Invasive prenatal diagnostic Obstetrics and Gynaecology, Queen Elizabeth Hospital, Gascoigne Road, techniques are indicated to exclude chromosomal abnormalities. Molecular analysis for some Kowloon, Hong Kong SAR, China syndromes is feasible if the family history is suggestive. Tel. +852-3506 6398 Fax. +852-2384 5834 E-mail: [email protected] Keywords: Craniofacial; Prenatal; Ultrasound; Three-dimensional ultrasonography; Fetal structural abnormalities This is an Open Access article distributed under the Introduction terms of the Creative Commons Attribution Non- Commercial License (http://creativecommons.org/ licenses/by-nc/3.0/) which permits unrestricted non- Craniofacial abnormalities are common. -

Identifying the Misshapen Head: Craniosynostosis and Related Disorders Mark S
CLINICAL REPORT Guidance for the Clinician in Rendering Pediatric Care Identifying the Misshapen Head: Craniosynostosis and Related Disorders Mark S. Dias, MD, FAAP, FAANS,a Thomas Samson, MD, FAAP,b Elias B. Rizk, MD, FAAP, FAANS,a Lance S. Governale, MD, FAAP, FAANS,c Joan T. Richtsmeier, PhD,d SECTION ON NEUROLOGIC SURGERY, SECTION ON PLASTIC AND RECONSTRUCTIVE SURGERY Pediatric care providers, pediatricians, pediatric subspecialty physicians, and abstract other health care providers should be able to recognize children with abnormal head shapes that occur as a result of both synostotic and aSection of Pediatric Neurosurgery, Department of Neurosurgery and deformational processes. The purpose of this clinical report is to review the bDivision of Plastic Surgery, Department of Surgery, College of characteristic head shape changes, as well as secondary craniofacial Medicine and dDepartment of Anthropology, College of the Liberal Arts characteristics, that occur in the setting of the various primary and Huck Institutes of the Life Sciences, Pennsylvania State University, State College, Pennsylvania; and cLillian S. Wells Department of craniosynostoses and deformations. As an introduction, the physiology and Neurosurgery, College of Medicine, University of Florida, Gainesville, genetics of skull growth as well as the pathophysiology underlying Florida craniosynostosis are reviewed. This is followed by a description of each type of Clinical reports from the American Academy of Pediatrics benefit from primary craniosynostosis (metopic, unicoronal, bicoronal, sagittal, lambdoid, expertise and resources of liaisons and internal (AAP) and external reviewers. However, clinical reports from the American Academy of and frontosphenoidal) and their resultant head shape changes, with an Pediatrics may not reflect the views of the liaisons or the emphasis on differentiating conditions that require surgical correction from organizations or government agencies that they represent. -

The Genetics of Canine Skull Shape Variation
PERSPECTIVES The Genetics of Canine Skull Shape Variation Jeffrey J. Schoenebeck and Elaine A. Ostrander1 Cancer Genetics Branch, National Human Genome Research Institute, National Institutes of Health, Bethesda, Maryland 20892 ABSTRACT A dog’s craniofacial diversity is the result of continual human intervention in natural selection, a process that began tens of thousands of years ago. To date, we know little of the genetic underpinnings and developmental mechanisms that make dog skulls so morphologically plastic. In this Perspectives, we discuss the origins of dog skull shapes in terms of history and biology and highlight recent advances in understanding the genetics of canine skull shapes. Of particular interest are those molecular genetic changes that are associated with the development of distinct breeds. OMETIME during the Paleolithic, a remarkable transfor- Variation in Skull Shape and Dog Domestication mation occurred. Small numbers of gray wolves adopted S Molecular clock estimates from mitochondrial DNA suggest a new pack master—humans. Through the process of do- domestication started as early as 135,000 years ago (Vilà mestication, the modern dog emerged. Today most dogs et al. 1997). More conservative estimates are based on ar- share little resemblance to their lupine ancestors. As a result chaeological records, which indicate that dog domestication of artificial selection, dogs radiated to fill niches in our lives, began somewhere between 15,000 and 36,000 years ago becoming our herders, guardians, hunters, rescuers, and com- (see summary by Larson et al. 2012). Current archaeologi- panions (Wilcox and Walkowicz 1995). The range of sizes cal estimates depend on carbon dating of bones, whose among dogs extends beyond that of wolves, giving dogs the morphologies appear distinct from that of contemporary distinction of being the most morphologically diverse terres- wolves. -

Review Article Cleidocranial Dysplasia: Clinical and Molecular Genetics
J Med Genet 1999;36:177–182 177 Review article Cleidocranial dysplasia: clinical and molecular genetics Stefan Mundlos Abstract Chinese named Arnold, was probably de- Cleidocranial dysplasia (CCD) (MIM scribed by Jackson.6 He was able to trace 356 119600) is an autosomal dominant skeletal members of this family of whom 70 were dysplasia characterised by abnormal aVected with the “Arnold Head”. CCD was clavicles, patent sutures and fontanelles, originally thought to involve only bones of supernumerary teeth, short stature, and a membranous origin. More recent and detailed variety of other skeletal changes. The dis- clinical investigations have shown that CCD is ease gene has been mapped to chromo- a generalised skeletal dysplasia aVecting not some 6p21 within a region containing only the clavicles and the skull but the entire CBFA1, a member of the runt family of skeleton. CCD was therefore considered to be transcription factors. Mutations in the a dysplasia rather than a dysostosis.7 Skeletal CBFA1 gene that presumably lead to syn- abnormalities commonly found include cla- thesis of an inactive gene product were vicular aplasia/hypoplasia, bell shaped thorax, identified in patients with CCD. The func- enlarged calvaria with frontal bossing and open tion of CBFA1 during skeletal develop- fontanelles, Wormian bones, brachydactyly ment was further elucidated by the with hypoplastic distal phalanges, hypoplasia of generation of mutated mice in which the the pelvis with widened symphysis pubis, Cbfa1 gene locus was targeted. Loss of one severe dental anomalies, and short stature. The Cbfa1 allele (+/-) leads to a phenotype very changes suggest that the gene responsible is not similar to human CCD, featuring hypo- only active during early development, as plasia of the clavicles and patent fonta- implied by changes in the shape or number of nelles. -

Cranial Sutures & Funny Shaped Heads: Radiological Diagnosis
Objectives • The objectives of this presentation are to: – Review the imaging features of normal cranial sutures – Identify the characteristics of abnormal skull shape on imaging – Review the characteristics of the most common non- syndromic and syndromic causes of craniosynostosis Anatomical Review Anatomical Review • The bony plates of the skull communicate at the cranial sutures • The anterior fontanelle occurs where the coronal & metopic sutures meet • The posterior fontanelle occurs where the sagittal & lambdoid sutures meet Anatomical Review • The main cranial sutures & fontanelles include: Metopic Suture Anterior Fontanelle Coronal Sutures Squamosal Sutures Posterior Fontanelle Sagittal Suture Lambdoid Sutures Anatomical Review • Growth of the skull occurs perpendicular to the cranial suture • This is controlled by a complex signalling system including: – Ephrins (mark the suture boundary) – Fibroblast growth factor receptors (FGFR) – Transcription factor TWIST Anatomical Review • The cranial sutures are important for rapid skull growth in-utero & infancy • The cranial sutures can usually be visualised on imaging into late adulthood Normal Radiological Appearances Normal Radiological Appearances • The cranial sutures can be visualised on plain radiographs • Standard views include: – PA – Lateral – Townes PA Skull radiograph Sagittal Suture Left Coronal Suture Right CoronalRight lambdoidSutureSuture Metopic Suture Left lambdoid Suture Townes View Sagittal Suture Left Coronal Suture Right Coronal Suture Right Lambdoid Suture Left -

A Heads up on Craniosynostosis
Volume 1, Issue 1 A HEADS UP ON CRANIOSYNOSTOSIS Andrew Reisner, M.D., William R. Boydston, M.D., Ph.D., Barun Brahma, M.D., Joshua Chern, M.D., Ph.D., David Wrubel, M.D. Craniosynostosis, an early closure of the growth plates of the skull, results in a skull deformity and may result in neurologic compromise. Craniosynostosis is surprisingly common, occurring in one in 2,100 children. It may occur as an isolated abnormality, as a part of a syndrome or secondary to a systemic disorder. Typically, premature closure of a suture results in a characteristic cranial deformity easily recognized by a trained observer. it is usually the misshapen head that brings the child to receive medical attention and mandates treatment. This issue of Neuro Update will present the diagnostic features of the common types of craniosynostosis to facilitate recognition by the primary care provider. The distinguishing features of other conditions commonly confused with craniosynostosis, such as plagiocephaly, benign subdural hygromas of infancy and microcephaly, will be discussed. Treatment options for craniosynostosis will be reserved for a later issue of Neuro Update. Types of Craniosynostosis Sagittal synostosis The sagittal suture runs from the anterior to the posterior fontanella (Figure 1). Like the other sutures, it allows the cranial bones to overlap during birth, facilitating delivery. Subsequently, this suture is the separation between the parietal bones, allowing lateral growth of the midportion of the skull. Early closure of the sagittal suture restricts lateral growth and creates a head that is narrow from ear to ear. However, a single suture synostosis causes deformity of the entire calvarium. -

Deformational Plagiocephaly, Brachycephaly, and Scaphocephaly
ORIGINAL ARTICLE Deformational Plagiocephaly, Brachycephaly, and Scaphocephaly. Part I: Terminology, Diagnosis, and Etiopathogenesis Gary F. Rogers, MD, JD, MBA, MPH oblique cranial length ratio of 106% or greater, and brachycephaly as Abstract: Cranial deformation is the most common cause of ab- a cephalic index (CI = cranial width/length) of 93% or greater. Using normal head shape. Intentional and unintentional alterations of these definitions, the combined prevalence of flattening was as high cranial form are associated with the application of external pressure as 19.7% at age 4 months and declined to 3.3% by 2 years of age. to the growing infant head, and such changes have been recorded However, when flattening was defined as an oblique cranial length throughout man’s history. Recent changes in Western sleeping ratio of 105% or greater and CI of 91% or greater, the prevalence practices, instituted to reduce the incidence of sudden infant death increased to 28% at 4 months and 12.7% at 2 years. There has been syndrome, have led to a dramatic rise in the incidence of cranial even more variation in the methods used to measure and report deformation and renewed interest in this subject. This 2-part review flattening, as discussed below. presents a pragmatic clinical approach to this topic including a critical review of the literature as it applies to each aspect of this DIAGNOSIS AND TERMINOLOGY common diagnosis: historical perspective, terminology, differential diagnosis, etiopathogenesis and predisposing factors, and prevention Deformational Plagiocephaly and treatment. Deformational cranial flattening can take many forms, de- pending on the position of the infant’shead during the first few months Key Words: Deformational plagiocephaly, deformational of life. -

EUROCAT Syndrome Guide
JRC - Central Registry european surveillance of congenital anomalies EUROCAT Syndrome Guide Definition and Coding of Syndromes Version July 2017 Revised in 2016 by Ingeborg Barisic, approved by the Coding & Classification Committee in 2017: Ester Garne, Diana Wellesley, David Tucker, Jorieke Bergman and Ingeborg Barisic Revised 2008 by Ingeborg Barisic, Helen Dolk and Ester Garne and discussed and approved by the Coding & Classification Committee 2008: Elisa Calzolari, Diana Wellesley, David Tucker, Ingeborg Barisic, Ester Garne The list of syndromes contained in the previous EUROCAT “Guide to the Coding of Eponyms and Syndromes” (Josephine Weatherall, 1979) was revised by Ingeborg Barisic, Helen Dolk, Ester Garne, Claude Stoll and Diana Wellesley at a meeting in London in November 2003. Approved by the members EUROCAT Coding & Classification Committee 2004: Ingeborg Barisic, Elisa Calzolari, Ester Garne, Annukka Ritvanen, Claude Stoll, Diana Wellesley 1 TABLE OF CONTENTS Introduction and Definitions 6 Coding Notes and Explanation of Guide 10 List of conditions to be coded in the syndrome field 13 List of conditions which should not be coded as syndromes 14 Syndromes – monogenic or unknown etiology Aarskog syndrome 18 Acrocephalopolysyndactyly (all types) 19 Alagille syndrome 20 Alport syndrome 21 Angelman syndrome 22 Aniridia-Wilms tumor syndrome, WAGR 23 Apert syndrome 24 Bardet-Biedl syndrome 25 Beckwith-Wiedemann syndrome (EMG syndrome) 26 Blepharophimosis-ptosis syndrome 28 Branchiootorenal syndrome (Melnick-Fraser syndrome) 29 CHARGE -
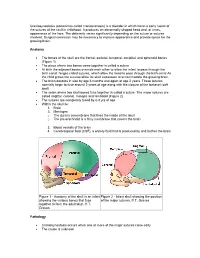
Craniosynostosis (Sometimes Called Craniostenosis) Is a Disorder in Which There Is Early Fusion of the Sutures of the Skull in Childhood
Craniosynostosis (sometimes called craniostenosis) is a disorder in which there is early fusion of the sutures of the skull in childhood. It produces an abnormally shaped head and, at times, appearance of the face. The deformity varies significantly depending on the suture or sutures involved. Surgical correction may be necessary to improve appearance and provide space for the growing brain. Anatomy • The bones of the skull are the frontal, parietal, temporal, occipital, and sphenoid bones (Figure 1) • The place where two bones come together is called a suture • At birth the adjacent bones override each other to allow the infant to pass through the birth canal. hinges called sutures, which allow the head to pass through the birth canal As the child grows the sutures allow for skull expansion to accommodate the growing brain. • The brain doubles in size by age 6 months and again at age 2 years. These sutures normally begin to fuse around 2 years of age along with the closure of the fontanel (soft spot) • The seam where two skull bones fuse together is called a suture. The major sutures are called sagittal, coronal, metopic and lambdoid (Figure 2) • The sutures are completely fused by 6-8 yrs of age • Within the skull lie: 1. Brain 2. Meninges o The dura is a membrane that lines the inside of the skull o The pia-arachnoid is a filmy membrane that covers the brain 3. Blood vessels of the brain 4. Cerebrospinal fluid (CSF), a watery fluid that is produced by and bathes the brain Figure 1 - Anatomy of the skull in an infant Figure 2 - Infant skull showing the position showing the various bones that fuse of the major sutures. -

Crouzon Syndrome
Other leaflets available from Headlines Craniofacial Support 6 Please contact Group Administrator Justine Tweedie - [email protected] for details of how to obtain copies 1 What causes Craniosynostosis? - a general discussion with a focus on genetic aspects (syndromes) 2 Apert Syndrome 3 Non-syndromic Craniosynostosis Headlines 4 Craniofacial Surgery 5 The Surgical Treatment of Hand Anomalies associated with Craniofacial Support Craniofacial conditions 6 Crouzon Syndrome 7 Pfeiffer Syndrome 8 Saethre-Chotzen Syndrome 11 Glossary of Terms associated with Craniosynostosis Crouzon Syndrome 12 Coping with Facial Disfigurement 13 The Genetic Background to Craniosynostosis 14 Breathing Problems in Craniofacial Syndromes 15 Muenke Syndrome 16 Eye Aspects of Craniofacial Conditions 17 Occipital (Positional) Plagiocephaly 18 Craniofrontonasal Syndrome Headlines - The Craniofacial Support Group Reg Name Headlines—The Craniofacial Support Group 1995 Reg Address 13 Heol Pentre’r Felin, Llantwit Major, Vale of Glamorgan, CF61 2XS Reg Charity No 1058461 Reg Charity No 1058461 www.headlines.org.uk Ref: HL6 Operation Age Indication Cranioplasty Infancy Skull expansion and remodelling, for cosmetic benefit and to relieve Shunt surgery Childhood Neurosurgical operation to reduce intracranial pressure Facial Childhood To protect the eyes, protect advancement Adolescence against breathing difficulty, and provide cosmetic benefit. Often preceded and followed by a Choanal dilation Childhood ENT procedures to improve the Grommet insertion airway and treat chronic ear Bone-anchored infection and hearing hearing aid Squint surgery Childhood To correct ocular squint and Tarsorrhaphy improve vision. Tarsorrhaphy may be used to protect against exposure damage to surface of eye. Further reading Assessments and Treatment of Craniosynostosis. Thompson D, Jones B M, Hayward R D, Harkness W. -

Salter-Harris Iii Fractures
HIGH YIELD PEDIATRIC BOARD EXAM BRANDON GREEN DO CHIEF OF PEDIATRIC ORTHOPEDICS NISWONGER CHILDRENS HOSPITAL ASSISTANT PROFESSOR EAST TENNESSEE STATE UNIVERSITY ORTHOPEDIC SURGERY RESIDENCY OBJECTIVES The following are the most commonly tested We will cover trauma, deformity, and Pediatric Orthopedic topics developmental issues Key words are still key…. FGFR-3? The most common skeletal dysplasia Mutation affects a single protein in fibroblast growth factor receptor-3 (FGFR- 3) gene, changing glycine to arginine at position 380. Result is growth retardation of the proliferative zone of the growth plate, resulting in short limbs. ACHONDROPLASIA The growth plates with the most growth (proximal humerus/distal femur) are most affected, resulting in rhizomelic (proximal more than distal) short stature. Extremities • trident hands (inability to approximate extended middle and ring finger) • bowed legs • radial head subluxation • muscular hypotonia Spine • thoracolumbar kyphosis- often resolves by walking age • short pedicles and decreased interpedicular distance from L1-S1 COMP? PSEUDOACHONDROPLASIA • SHORT-LIMBED RHIZOMELIC DWARFISM WITH NORMAL FACIAL FEATURES • AUTOSOMAL DOMINANT • MUTATION IS IN CARTILAGE OLIGOMERIC MATRIX PROTEIN (COMP) ON CHROMOSOME 19. • EPIPHYSES ARE DELAYED AND ABNORMAL, METAPHYSEAL FLARING IS PRESENT, AND EARLY ONSET OSTEOARTHRITIS (OA) IS COMMON. SULFATE TRANSPORT GENE??? DIASTROPHIC DYSPLASIA • AUTOSOMAL RECESSIVE • MUTATION IN SULFATE TRANSPORT PROTEIN THAT PRIMARILY AFFECTS CARTILAGE MATRIX. DIASTROPHIC DYSPLASIA