30 August 2021 Aperto
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Chapter 11 Marine Fungi Associated with Antarctic Macroalgae
Chapter 11 Marine Fungi Associated with Antarctic Macroalgae Mayara B. Ogaki, Maria T. de Paula, Daniele Ruas, Franciane M. Pellizzari, César X. García-Laviña, and Luiz H. Rosa Abstract Fungi are well known for their important roles in terrestrial ecosystems, but filamentous and yeast forms are also active components of microbial communi- ties from marine ecosystems. Marine fungi are particularly abundant and relevant in coastal systems where they can be found in association with large organic substrata, like seaweeds. Antarctica is a rather unexplored region of the planet that is being influenced by strong and rapid climate change. In the past decade, several efforts have been made to get a thorough inventory of marine fungi from different environ- ments, with a particular emphasis on those associated with the large communities of seaweeds that abound in littoral and infralittoral ecosystems. The algicolous fungal communities obtained were characterized by a few dominant species and a large number of singletons, as well as a balance among endemic, indigenous, and cold- adapted cosmopolitan species. The long-term monitoring of this balance and the dynamics of richness, dominance, and distributional patterns of these algicolous fungal communities is proposed to understand and model the influence of climate change on the maritime Antarctic biota. In addition, several fungal isolates from marine Antarctic environments have shown great potential as producers of bioactive natural products and enzymes and may represent attractive sources of biotechno- logical products. M. B. Ogaki · M. T. de Paula · D. Ruas · L. H. Rosa (*) Departamento de Microbiologia, Universidade Federal de Minas Gerais, Belo Horizonte, MG, Brazil e-mail: [email protected] F. -

Endophytic Fungi: Biological Control and Induced Resistance to Phytopathogens and Abiotic Stresses
pathogens Review Endophytic Fungi: Biological Control and Induced Resistance to Phytopathogens and Abiotic Stresses Daniele Cristina Fontana 1,† , Samuel de Paula 2,*,† , Abel Galon Torres 2 , Victor Hugo Moura de Souza 2 , Sérgio Florentino Pascholati 2 , Denise Schmidt 3 and Durval Dourado Neto 1 1 Department of Plant Production, Luiz de Queiroz College of Agriculture, University of São Paulo, Piracicaba 13418900, Brazil; [email protected] (D.C.F.); [email protected] (D.D.N.) 2 Plant Pathology Department, Luiz de Queiroz College of Agriculture, University of São Paulo, Piracicaba 13418900, Brazil; [email protected] (A.G.T.); [email protected] (V.H.M.d.S.); [email protected] (S.F.P.) 3 Department of Agronomy and Environmental Science, Frederico Westphalen Campus, Federal University of Santa Maria, Frederico Westphalen 98400000, Brazil; [email protected] * Correspondence: [email protected]; Tel.: +55-54-99646-9453 † These authors contributed equally to this work. Abstract: Plant diseases cause losses of approximately 16% globally. Thus, management measures must be implemented to mitigate losses and guarantee food production. In addition to traditional management measures, induced resistance and biological control have gained ground in agriculture due to their enormous potential. Endophytic fungi internally colonize plant tissues and have the potential to act as control agents, such as biological agents or elicitors in the process of induced resistance and in attenuating abiotic stresses. In this review, we list the mode of action of this group of Citation: Fontana, D.C.; de Paula, S.; microorganisms which can act in controlling plant diseases and describe several examples in which Torres, A.G.; de Souza, V.H.M.; endophytes were able to reduce the damage caused by pathogens and adverse conditions. -

New Taxa in Aspergillus Section Usti
available online at www.studiesinmycology.org StudieS in Mycology 69: 81–97. 2011. doi:10.3114/sim.2011.69.06 New taxa in Aspergillus section Usti R.A. Samson1*, J. Varga1,2, M. Meijer1 and J.C. Frisvad3 1CBS-KNAW Fungal Biodiversity Centre, Uppsalalaan 8, NL-3584 CT Utrecht, the Netherlands; 2Department of Microbiology, Faculty of Science and Informatics, University of Szeged, H-6726 Szeged, Közép fasor 52, Hungary; 3BioCentrum-DTU, Building 221, Technical University of Denmark, DK-2800 Kgs. Lyngby, Denmark. *Correspondence: Robert A. Samson, [email protected] Abstract: Based on phylogenetic analysis of sequence data, Aspergillus section Usti includes 21 species, inclucing two teleomorphic species Aspergillus heterothallicus (= Emericella heterothallica) and Fennellia monodii. Aspergillus germanicus sp. nov. was isolated from indoor air in Germany. This species has identical ITS sequences with A. insuetus CBS 119.27, but is clearly distinct from that species based on β-tubulin and calmodulin sequence data. This species is unable to grow at 37 °C, similarly to A. keveii and A. insuetus. Aspergillus carlsbadensis sp. nov. was isolated from the Carlsbad Caverns National Park in New Mexico. This taxon is related to, but distinct from a clade including A. calidoustus, A. pseudodeflectus, A. insuetus and A. keveii on all trees. This species is also unable to grow at 37 °C, and acid production was not observed on CREA. Aspergillus californicus sp. nov. is proposed for an isolate from chamise chaparral (Adenostoma fasciculatum) in California. It is related to a clade including A. subsessilis and A. kassunensis on all trees. This species grew well at 37 °C, and acid production was not observed on CREA. -

Diversity and Co-Occurrence Patterns of Microbial Communities in the Fertilized Eggs of Thitarodes Insect, the Implication on the Occurrence of Chinese Cordyceps
Diversity and Co-Occurrence Patterns of microbial Communities in the Fertilized Eggs of Thitarodes Insect, the Implication on the Occurrence of Chinese cordyceps Lian-Xian Guo Guangdong Medical University Xiao-Shan Liu Guangdong Medical University Zhan-Hua Mai Guangdong Medical University Yue-Hui Hong Guangdong Hospital of Traditional Chinese Medicine Qi-Jiong Zhu Guangdong Medical University Xu-Ming Guo Guangdong Medical University Huan-Wen Tang ( [email protected] ) Guangdong Medical University Research article Keywords: Bacterial, Fungal, Fertilized Eggs, Thitarodes, Chinese cordyceps, Wolbachia Posted Date: April 28th, 2020 DOI: https://doi.org/10.21203/rs.3.rs-24686/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/20 Abstract Background The large-scale articial cultivation of Chinese cordyceps has not been widely implemented because the crucial factors triggering the occurrence of Chinese cordyceps have not been fully illuminated. Methods In this study, the bacterial and fungal structure of fertilized eggs in the host Thitarodes collected from 3 sampling sites with different occurrence rates of Chinese cordyceps (Sites A, B and C: high, low and null Chinese cordyceps, respectively) were analyzed by performing 16S RNA and ITS sequencing, respectively. And the intra-kingdom and inter-kingdom network were analyzed. Results For bacterial community, totally 4671 bacterial OTUs were obtained. α-diversity analysis revealed that the evenness of the eggs from site A was signicantly higher than that of sites B and C, and the dominance index of site A was signicantly lower than that of sites B and C ( P < 0.05). -

Tesis (4.391Mb)
UNIVERSIDAD AUTÓNOMA DEL ESTADO DE MORELOS INSTITUTO DE INVESTIGACIÓN EN CIENCIAS BÁSICAS Y APLICADAS CENTRO DE INVESTIGACIÓN EN DINÁMICA CELULAR Análisis morfológico y molecular de Aspergillus sydowii en condiciones de baja actividad de agua. T E S I S QUE PARA OBTENER EL GRADO DE MAESTRO EN CIENCIAS PRESENTA: Lic. Gisell Valdés Muñoz DIRECTOR DE TESIS Dr. Ramón Alberto Batista García Sinodales Cuernavaca, Morelos Enero, 2021 Sinodales 1 Sinodales Dr. Carlos Eliud Angulo Valadez Dra. Sonia Dávila Ramos Dr. Raúl Peralta Rodríguez Dra. María del Rayo Sánchez Carbente Dr. Ramón Alberto Batista García 2 3 4 Agradecimientos Estos dos años de maestría en México han sido de los mejores de mi vida, he aprendido tantas cosas nuevas y he superado muchos retos incluso miedos, y hay tantas personas a las cuales agradecer: A mi tutor Dr. Ramón A. Batista García, por su aceptación, guía, palabras fuertes y cariñosas, y por toda su destreza para elaborar un camino bonito de estudios y superación para mí. A mi asesora y amiga MsC. Irina Jiménez Gómez, por cada día enseñarme algo nuevo, por enseñarme a trabajar en el laboratorio con los hongos, por nuestras experiencias microscópicas en Ensenada y porque verdaderamente con ella mi maestría fue excelente. A la Dra. María del Rayo Sánchez Carbente y Dra. Sonia Dávila Ramos porque en cada examen semestral me apoyaron a mejorar esta investigación. Al Dr. Ayixon Sánchez Reyes, MsC. Hugo Castelán y MsC. Yordanis Pérez Llano por su ayuda bioinformática tan amable y desinteresada, gracias por compartir esos conocimientos. A las chicas ―Aspergillus‖ Debu, Heidy, Lyselle, Adri por nuestros estudios en conjunto, por darnos apoyo y siempre estar disponibles, gracias por su amistad. -

Lists of Names in Aspergillus and Teleomorphs As Proposed by Pitt and Taylor, Mycologia, 106: 1051-1062, 2014 (Doi: 10.3852/14-0
Lists of names in Aspergillus and teleomorphs as proposed by Pitt and Taylor, Mycologia, 106: 1051-1062, 2014 (doi: 10.3852/14-060), based on retypification of Aspergillus with A. niger as type species John I. Pitt and John W. Taylor, CSIRO Food and Nutrition, North Ryde, NSW 2113, Australia and Dept of Plant and Microbial Biology, University of California, Berkeley, CA 94720-3102, USA Preamble The lists below set out the nomenclature of Aspergillus and its teleomorphs as they would become on acceptance of a proposal published by Pitt and Taylor (2014) to change the type species of Aspergillus from A. glaucus to A. niger. The central points of the proposal by Pitt and Taylor (2014) are that retypification of Aspergillus on A. niger will make the classification of fungi with Aspergillus anamorphs: i) reflect the great phenotypic diversity in sexual morphology, physiology and ecology of the clades whose species have Aspergillus anamorphs; ii) respect the phylogenetic relationship of these clades to each other and to Penicillium; and iii) preserve the name Aspergillus for the clade that contains the greatest number of economically important species. Specifically, of the 11 teleomorph genera associated with Aspergillus anamorphs, the proposal of Pitt and Taylor (2014) maintains the three major teleomorph genera – Eurotium, Neosartorya and Emericella – together with Chaetosartorya, Hemicarpenteles, Sclerocleista and Warcupiella. Aspergillus is maintained for the important species used industrially and for manufacture of fermented foods, together with all species producing major mycotoxins. The teleomorph genera Fennellia, Petromyces, Neocarpenteles and Neopetromyces are synonymised with Aspergillus. The lists below are based on the List of “Names in Current Use” developed by Pitt and Samson (1993) and those listed in MycoBank (www.MycoBank.org), plus extensive scrutiny of papers publishing new species of Aspergillus and associated teleomorph genera as collected in Index of Fungi (1992-2104). -

Parkinsonia Aculeata
Investigating the cause of dieback in the invasive plant, Parkinsonia aculeata BY TRACEY VIVIEN STEINRUCKEN A thesis submitted in fulfilment of the requirements for the degree of Doctor of Philosophy at Western Sydney University in 2017 This page has been intentionally left blank “Watch with glittering eyes the whole world around you because the greatest secrets are always hidden in the most unlikely places. Those who don't believe in magic will never find it” -- Roald Dahl This page has been intentionally left blank Acknowledgements I would like to thank my advisors Rieks van Klinken (CSIRO Health & Biosecurity), Andrew Bissett (CISRO Oceans & Atmosphere) and Jeff Powell (Hawkesbury Institute for the Enivronment, Western Sydney University) for their excellent mentoring, patient communication across borders and constant support. This research project was supported by Meat and Livestock Australia via a technical assistance grant (B.STU.0271). My PhD was supported by the Australian Government via an Australian Postgraduate Award and Western Sydney University via a top-up stipend. The Hawkesbury Institute for the Environment also supported my work with an annual research allocation and conference attendance funding. Thanks to Patricia Hellier, David Harland, Ian Anderson and Lisa Davison at HIE for administrative support. Thank-you to Kelli Pukallus (Biosecurity Queensland), Andrew White (CSIRO), Eva Pôtet (Agro Campus Oest, Paris), Marcus Klein (HIE at WSU), Donald Gardiner (CSIRO), Shamsul Hoque (CSIRO), Ryan O’Dell (DAFF) and Dylan Smith (UC Berkeley) for field and technical support in various chapters throughout this thesis. Huge thanks to my CSIRO Biosecurity team: Gio Fichera, Ryan Zonneveld, Brad Brown, Andrew White and Jeff Makinson for technical support in Chapter 3. -

Secondary Metabolites from Eurotium Species, Aspergillus Calidoustus and A
mycological research 113 (2009) 480–490 journal homepage: www.elsevier.com/locate/mycres Secondary metabolites from Eurotium species, Aspergillus calidoustus and A. insuetus common in Canadian homes with a review of their chemistry and biological activities Gregory J. SLACKa, Eva PUNIANIa, Jens C. FRISVADb, Robert A. SAMSONc, J. David MILLERa,* aOttawa-Carleton Institute of Chemistry, Carleton University, Ottawa, ON, Canada K1S 5B6 bDepartment of Systems Biology, Technical University of Denmark, DK-2800 Lyngby, Denmark cCBS Fungal Biodiversity Centre, PO Box 85167, NL-3508 AD Utrecht, The Netherlands article info abstract Article history: As part of studies of metabolites from fungi common in the built environment in Canadian Received 22 July 2008 homes, we investigated metabolites from strains of three Eurotium species, namely Received in revised form E. herbariorum, E. amstelodami, and E. rubrum as well as a number of isolates provisionally 26 November 2008 identified as Aspergillus ustus. The latter have been recently assigned as the new species Accepted 16 December 2008 A. insuetus and A. calidoustus. E. amstelodami produced neoechinulin A and neoechinulin Published online 14 January 2009 B, epiheveadride, flavoglaucin, auroglaucin, and isotetrahydroauroglaucin as major metab- Corresponding Editor: olites. Minor metabolites included echinulin, preechinulin and neoechinulin E. E. rubrum Stephen W. Peterson produced all of these metabolites, but epiheveadride was detected as a minor metabolite. E. herbariorum produced cladosporin as a major metabolite, in addition to those found in Keywords: E. amstelodami. This species also produced questin and neoechinulin E as minor metabo- Aspergillus insuetus lites. This is the first report of epiheveadride occurring as a natural product, and the first A. -

{Replace with the Title of Your Dissertation}
Mechanisms of Invasion and the Microbiome of Introduced Species A Dissertation SUBMITTED TO THE FACULTY OF UNIVERSITY OF MINNESOTA BY Aaron S. David IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY Eric Seabloom Georgiana May May 2016 © Aaron S. David 2016 Acknowledgements I have been fortunate to have had incredible guidance, mentorship, and assistance throughout my time as a Ph.D. student at the University of Minnesota. I would like to start by acknowledging and thanking my advisors, Dr. Eric Seabloom and Dr. Georgiana May for providing crucial support, and always engaging me in stimulating discussion. I also thank my committee members, Dr. Peter Kennedy, Dr. Linda Kinkel, and Dr. David Tilman for their guidance and expertise. I am indebted to Dr. Sally Hacker and Dr. Joey Spatafora of Oregon State University for generously welcoming me into their laboratories while I conducted my field work. Dr. Phoebe Zarnetske and Shawn Gerrity showed me the ropes out on the dunes and provided valuable insight along the way. I also have to thank the many undergraduate students who helped me in the field in laboratory. In particular, I need to thank Derek Schmidt, who traveled to Oregon with me and helped make my field work successful. I also thank my other collaborators that made this work possible, especially Dr. Peter Ruggiero and Reuben Biel who contributed to the data collection and analysis in Chapter 1, and Dr. Gina Quiram and Jennie Sirota who contributed to the study design and data collection in Chapter 4. I would also like to thank the amazing faculty, staff, and students of Ecology, Evolution, and Behavior and neighboring departments. -

Epidemiology, Treatment Options and Outcome of Invasive Infections Caused by Aspergillus Section Usti
Unicentre CH-1015 Lausanne http://serval.unil.ch Year : 2020 Epidemiology, treatment options and outcome of invasive infections caused by Aspergillus section Usti Glampedakis Emmanouil Glampedakis Emmanouil, 2020, Epidemiology, treatment options and outcome of invasive infections caused by Aspergillus section Usti Originally published at : Thesis, University of Lausanne Posted at the University of Lausanne Open Archive http://serval.unil.ch Document URN : urn:nbn:ch:serval-BIB_D909780962850 Droits d’auteur L'Université de Lausanne attire expressément l'attention des utilisateurs sur le fait que tous les documents publiés dans l'Archive SERVAL sont protégés par le droit d'auteur, conformément à la loi fédérale sur le droit d'auteur et les droits voisins (LDA). A ce titre, il est indispensable d'obtenir le consentement préalable de l'auteur et/ou de l’éditeur avant toute utilisation d'une oeuvre ou d'une partie d'une oeuvre ne relevant pas d'une utilisation à des fins personnelles au sens de la LDA (art. 19, al. 1 lettre a). A défaut, tout contrevenant s'expose aux sanctions prévues par cette loi. Nous déclinons toute responsabilité en la matière. Copyright The University of Lausanne expressly draws the attention of users to the fact that all documents published in the SERVAL Archive are protected by copyright in accordance with federal law on copyright and similar rights (LDA). Accordingly it is indispensable to obtain prior consent from the author and/or publisher before any use of a work or part of a work for purposes other than personal use within the meaning of LDA (art. -

Corrigiendo Tesis Doctorado Paloma Casas Junco
TECNOLÓGICO NACIONAL DE MÉXICO Instituto Tecnológico de Tepic EFECTO DE PLASMA FRÍO EN LA REDUCCIÓN DE OCRATOXINA A EN CAFÉ DE NAYARIT (MÉXICO) TESIS Por: MCA. PALOMA PATRICIA CASAS JUNCO DOCTORADO EN CIENCIAS EN ALIMENTOS Director: Dra. Montserrat Calderón Santoyo Co - director: Dr. Juan Arturo Ragazzo Sánchez Tepic, Nayarit Febrero 2018 RESUMEN Casas-Junco, Paloma Patricia. DCA. Instituto Tecnológico de Tepic. Febrero de 2018. Efecto de plasma frío en la reducción de ocratoxina A en café de Nayarit (México). Directora: Montserrat Calderón Santoyo. La ocratoxina A (OTA) se considera uno de los principales problemas emergentes en la industria del café, dado que el proceso de tostado no asegura su destrucción total. El objetivo de este estudio fue identificar las especies fúngicas productoras de OTA en café tostado de Nayarit, así como evaluar el efecto de plasma frío en la inhibición de esporas de hongos micotoxigénicos, detoxificación de OTA, así como en algunos parámetros de calidad del café. Se aislaron e identificaron hongos micotoxigénicos mediante claves dicotómicas, después se analizó la producción de OTA y aflatoxinas (AFB1, AFB2, AFG2, AFG1) por HPLC con detector de fluorescencia. Las cepas productoras de toxinas se identificaron por PCR utilizando los primers ITS1 e ITS4. Después se aplicó plasma frío en muestras de café tostado inoculadas con hongos micotoxigénicos (A. westerdijikiae, A. steynii, A. niger y A. versicolor) a diferentes tiempos 0, 1, 2, 4, 5, 6, 8, 10, 12, 14, 16 y 18 min, con una potencia de entrada 30 W y un voltaje de salida de 850 voltios y helio publicitario (1.5 L/min). -
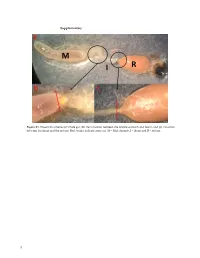
Supplementary File 1
Supplementary Figure S1. Dissection scheme (a) whole gut, (b) the transition between the middle stomach and ileum, and (c) transition between the ileum and the rectum. Red streaks indicate areas cut. M = Mid stomach, I = ileum and R = rectum. 1 Figure S2. Relative abundance of the 20 most abundant fungal OTUs across months per gut part. Taxonomic assignments are provided in Table S1. 2 Figure S3. Relative abundance of the bacterial OUTs (with abundance > 1%) across months per gut part. The color code is given at the genus level. 3 Table S1. Taxonomic assignments of fungal OTUs by Blast. OTU# Taxonomy by blast nt collection 11/11‐2020 Phylum Accesion# OTU_1 Aureobasidium pullulans Ascomycota MW085051 OTU_2 Meyerozyma guilliermondii Ascomycota MT988167 OTU_3 Starmerella apicola Ascomycota KY101940 OTU_4 Debaryomyces hansenii Ascomycota MW051606 OTU_5 Engyodontium sp. Ascomycota MN905797 Basidiomy‐ OTU_6 Mrakia sp. MT505696 cota Basidiomy‐ OTU_7 Tausonia pullulans MN900123 cota OTU_8 Unknown OTU_9 Unknown OTU_10 Penicillium corylophilum Ascomycota MT906500 OTU_11 Penicillium sp. Ascomycota MT993349 OTU_12 Cladosporium sp. Ascomycota MW077705 Phaffia rhodozyma/Xanthophyllomyces dendro‐ Basidiomy‐ OTU_13 KY104501/DQ904243 rhous cota Basidiomy‐ OTU_14 Filobasidium wieringae MN899199 cota OTU_15 Hanseniaspora uvarum Ascomycota MN556596 Basidiomy‐ OTU_16 Mrakia gelida MN460370 cota OTU_17 Uncultured fungus clone MK717974 OTU_18 Cladosporium allicinum Ascomycota MT974153 OTU_20 Taphrina carpini Ascomycota MK782181 Basidiomy‐ OTU_25 Papiliotrema