Genetic Admixture Analysis in the Population of Tacuarembó Uruguay Using Alu Insertions
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

U R U G U a Y
Telephone: 598 26040329 U R U G U A Y ext. 1260, 1463 Dirección Nacional de Aviación Civil e Infraestructura Aeronáutica AMDT Telefax: 598 26040067 Servicio de Información Aeronáutica NR 56 AFTN: SUMUYNYX Aeropuerto Intl de Carrasco “Gral. Cesáreo L. Berisso” 01 DEC 2018 e-mail: [email protected] 14000 Canelones The entries with an indicator (!) at the margin indicates changes in the paragraph EFFECTIVE DATE: 01 DEC 2018 - 03:01 UTC THIS AMDT MUST NOT BE INSERTED INTO THE AIP BEFORE THE EFFECTIVE DATE. HOWEVER, IT IS SUGGESTED TO STUDY ITS CONTENT BEFORE THAT DATE. 1. INSERT AND/OR DESTROY THE FOLLOWING PAGES: DESTROY INSERT GEN GEN 0.4-1 ........................................ 11 OCT 2018 0.4-1............................... 01 DEC 2018 0.4-2 ........................................ 11 OCT 2018 0.4-2............................... 01 DEC 2018 0.4-3 ........................................ 11 OCT 2018 0.4-3............................... 01 DEC 2018 0.4-4 ........................................ 01 APR 2018 0.4-4............................... 01 DEC 2018 0.4-5 ........................................ 01 AUG 2018 0.4-5............................... 01 DEC 2018 0.4-6 ........................................ 01 DEC 2017 1.5-1....... ................................. 02 JAN 2017 1.5-1............................... 01 DEC 2018 1.6-1 ........................................ 02 JAN 2017 1.6-1............................... 01 DEC 2018 1.6-2 ........................................ 01 APR 2002 1.6-3 ........................................ 01 APR 2002 1.6-4 ........................................ 01 APR 2014 1.6-5 ........................................ 01 DEC 2010 1.6-6 ........................................ 01 DEC 2010 1.6-7 ........................................ 01 APR 2014 1.6-8 ........................................ 01 APR 2014 1.6-9 ........................................ 01 APR 2014 1.6-10 ...................................... 02 JAN 2017 1.6-11 ...................................... 01 APR 2014 1.6-12 ..................................... -

Women, Business and the Law 2020 World Bank Group
WOMEN, BUSINESS AND THE LAW 2020 AND THE LAW BUSINESS WOMEN, WOMEN, BUSINESS AND THE LAW 2020 WORLD BANK GROUP WORLD WOMEN, BUSINESS AND THE LAW 2020 © 2020 International Bank for Reconstruction and Development / The World Bank 1818 H Street NW, Washington, DC 20433 Telephone: 202-473-1000; Internet: www.worldbank.org Some rights reserved 1 2 3 4 23 22 21 20 This work is a product of the staff of The World Bank with external contributions. The findings, interpretations, and conclusions expressed in this work do not necessarily reflect the views of The World Bank, its Board of Executive Directors, or the govern- ments they represent. The World Bank does not guarantee the accuracy of the data included in this work. The boundaries, colors, denominations, and other information shown on any map in this work do not imply any judgment on the part of The World Bank concerning the legal status of any territory or the endorsement or acceptance of such boundaries. Nothing herein shall constitute or be considered to be a limitation upon or waiver of the privileges and immunities of The World Bank, all of which are specifically reserved. Rights and Permissions This work is available under the Creative Commons Attribution 3.0 IGO license (CC BY 3.0 IGO) http://creativecommons.org/ licenses/by/3.0/igo. Under the Creative Commons Attribution license, you are free to copy, distribute, transmit, and adapt this work, including for commercial purposes, under the following conditions: Attribution—Please cite the work as follows: World Bank. 2020. Women, Business and the Law 2020. -

John Benjamins Publishing Company
John Benjamins Publishing Company This is a contribution from New Approaches to Language Attitudes in the Hispanic and Lusophone World. Edited by Talia Bugel and Cecilia Montes-Alcalá. © 2020. John Benjamins Publishing Company This electronic file may not be altered in any way. The author(s) of this article is/are permitted to use this PDF file to generate printed copies to be used by way of offprints, for their personal use only. Permission is granted by the publishers to post this file on a closed server which is accessible to members (students and staff) only of the author’s/s’ institute, it is not permitted to post this PDF on the open internet. For any other use of this material prior written permission should be obtained from the publishers or through the Copyright Clearance Center (for USA: www.copyright.com). Please contact [email protected] or consult our website: www.benjamins.com Tables of Contents, abstracts and guidelines are available at www.benjamins.com Attitudes toward Portuguese in Uruguay in the nineteenth century Virginia Bertolotti and Magdalena Coll Universidad de la República The Uruguay-Brazil border has been a porous geographical area since colonial times, giving rise to intense contact between Portuguese and Spanish, further fostered by a shared economic and demographic foundation. Here we analyze at- titudes toward border Portuguese and that language in general in the Uruguayan territory during the Luso-Brazilian military occupation (1816–1828) and later in the century and identify two parallel realities: a predominant neutral attitude toward Portuguese, as the language was not an object of evaluation; and the ex- istence of negative attitudes toward Portuguese promoted by the authorities in Montevideo. -

® Wiseburn Administrators Earn Honors Seeking the Truth of Ancestry
FREE Education + Communication = A Better Nation ® Covering the Wiseburn School District VOLUME 5, IssUE 13 www.SchoolNewsRollCall.com MARCh–MAY 2013 SUPERINTENDENT Wiseburn Administrators Earn Honors Wiseburn By Chris Jones, Ed.D. Director of Curriculum, Unification and Instruction, and Technology Wiseburn High Dr. Tom Johnstone: Pepperdine School Project University Superintendent of the Year Closing in on Since 2008, Superintendent Dr. Tom Johnston Success has led the Wiseburn School District toward As we move new levels of success. His boundless energy and deeper into 2013, determination to create innovative programs Dr. Tom the Wiseburn School Johnstone designed to help all students succeed have resulted District continues to in his being designated as Superintendent of forge ahead on two very significant the Year for 2013 by Pepperdine University. This fronts: Wiseburn unification and the distinction caps off a year of unprecedented Wiseburn High School Project. milestones achieved by Wiseburn in student There is much excitement in academic performance, facilities upgrades, and the community as Wiseburn gets significant progress in the long-desired movement closer to establishing a beautiful towards a unified K–12 Wiseburn School District. high school facility that will be Before joining the Wiseburn School District, Dr. the centerpiece of the Wiseburn Johnstone was a familiar face around town, as two community in the decades that of his children attended Anza School, where his lie ahead. On January 11, 2013, wife Terry is a first-grade teacher. Dr. Johnstone the district started the 45-day had established his professional reputation in the public review period for the Draft area by faithfully serving students in the Lennox Environmental Impact Report School District for 28 years as a teacher, counselor, (DEIR) for Wiseburn High School. -

Diaspora Engagement on Country Entrepreneurship and Investment Poli Cy Trends and Notable Practices in the Rabat Process Region
Background Paper Diaspora Engagement on Country Entrepreneurship and Investment Poli cy Trends and Notable Practices in the Rabat Process region Authors Valérie Wolff, International Centre for Migration Policy Development ICMPD Stella Opoku-Owusu, African Foundation for Development, AFFORD Diasmer Bloe, African Foundation for Development, AFFORD Background paper coordinated by the Secretariat of the Rabat Process, ICMPD Background paper for the Rabat Process Thematic Meeting on Diaspora Engagement Strategies: Entrepreneurship and Investment, 5-6 October in Bamako, Mali Project financed by the European Union Project implemented by ICMPD In the framework of the project “Support to Africa-EU Migration and Mobility Dialogue (MMD)” Introduction Diaspora entrepreneurship and investments are often hailed as drivers of economic development and positive change, and there is enough evidence to support this: 80% of FDI in China is from the Chinese diaspora, Indian diasporas in the USA have played an instrumental role in building up India’s IT industry and creating a second ‘Silicon Valley’ in their country -- to name some of the well-known success stories. This is no different to African diaspora actors. So who are these people and how have they become so engaged with their home countries? Often classified as ‘diaspora entrepreneurs’, they represent the group of migrants or those of migrant origin who are entrepreneurs, live outside of their home country, yet stay involved.1 It implies a transnational element of entrepreneurship between one or more -

Terrorist and Organized Crime Groups in the Tri-Border Area (Tba) of South America
TERRORIST AND ORGANIZED CRIME GROUPS IN THE TRI-BORDER AREA (TBA) OF SOUTH AMERICA A Report Prepared by the Federal Research Division, Library of Congress under an Interagency Agreement with the Crime and Narcotics Center Director of Central Intelligence July 2003 (Revised December 2010) Author: Rex Hudson Project Manager: Glenn Curtis Federal Research Division Library of Congress Washington, D.C. 205404840 Tel: 2027073900 Fax: 2027073920 E-Mail: [email protected] Homepage: http://loc.gov/rr/frd/ p 55 Years of Service to the Federal Government p 1948 – 2003 Library of Congress – Federal Research Division Tri-Border Area (TBA) PREFACE This report assesses the activities of organized crime groups, terrorist groups, and narcotics traffickers in general in the Tri-Border Area (TBA) of Argentina, Brazil, and Paraguay, focusing mainly on the period since 1999. Some of the related topics discussed, such as governmental and police corruption and anti–money-laundering laws, may also apply in part to the three TBA countries in general in addition to the TBA. This is unavoidable because the TBA cannot be discussed entirely as an isolated entity. Based entirely on open sources, this assessment has made extensive use of books, journal articles, and other reports available in the Library of Congress collections. It is based in part on the author’s earlier research paper entitled “Narcotics-Funded Terrorist/Extremist Groups in Latin America” (May 2002). It has also made extensive use of sources available on the Internet, including Argentine, Brazilian, and Paraguayan newspaper articles. One of the most relevant Spanish-language sources used for this assessment was Mariano César Bartolomé’s paper entitled Amenazas a la seguridad de los estados: La triple frontera como ‘área gris’ en el cono sur americano [Threats to the Security of States: The Triborder as a ‘Grey Area’ in the Southern Cone of South America] (2001). -

A •Œneo-Abolitionist Trendâ•Š in Sub-Saharan Africa? Regional Anti
Seattle Journal for Social Justice Volume 9 Issue 2 Spring/Summer 2011 Article 4 May 2011 A “Neo-Abolitionist Trend” in Sub-Saharan Africa? Regional Anti- Trafficking Patterns and a Preliminary Legislative Taxonomy Benjamin N. Lawrance Ruby P. Andrew Follow this and additional works at: https://digitalcommons.law.seattleu.edu/sjsj Recommended Citation Lawrance, Benjamin N. and Andrew, Ruby P. (2011) "A “Neo-Abolitionist Trend” in Sub-Saharan Africa? Regional Anti-Trafficking Patterns and a Preliminary Legislative Taxonomy," Seattle Journal for Social Justice: Vol. 9 : Iss. 2 , Article 4. Available at: https://digitalcommons.law.seattleu.edu/sjsj/vol9/iss2/4 This Article is brought to you for free and open access by the Student Publications and Programs at Seattle University School of Law Digital Commons. It has been accepted for inclusion in Seattle Journal for Social Justice by an authorized editor of Seattle University School of Law Digital Commons. For more information, please contact [email protected]. 599 A “Neo-Abolitionist Trend” in Sub-Saharan Africa? Regional Anti-Trafficking Patterns and a Preliminary Legislative Taxonomy Benjamin N. Lawrance and Ruby P Andrew INTRODUCTION In the decade since the US Department of State (USDOS) instituted the production of its annual Trafficking in Persons (TIP) Report, anti- trafficking measures have been passed across the globe, and the US role in setting standards and paradigms for anti-trafficking has been extensively analyzed and criticized.1 Through its Office to Monitor and Combat Trafficking in Persons (OMCTP), the USDOS’ three-tier categorization of nations’ trafficking measures continues to play a pivotal role in shaming recalcitrant nations into responding to accusations of trafficking within and across their respective borders.2 No region has experienced so dramatic a transformation with respect to the USDOS agenda of “prevention, protection, and prosecution” than sub- Saharan Africa.3 No sub-Saharan African nation had an anti-trafficking statute at the start of the millennium. -

Vulnerability of Family Livestock Farming on the Livramento-Rivera Border of Brazil and Uruguay: Comparative Analysis
Vulnerability of family livestock farming on the Livramento-Rivera border of Brazil and Uruguay: Comparative analysis Paulo D. Waquil1* Marcio Z. Neske1 Claudio M. Ribeiro2,3 Fabio E. Schlick2 Tanice Andreatta4 Cleiton Perleberg4 Marcos F.S. Borba5 Jose P. Trindade5 Rafael Carriquiry6,7 Italo Malaquin7 Alejandro Saravia7,8 Mario Gonzales9 Livio S.D. Claudino10 Keywords Summary Cattle, family farm, risk factor, Social, ecological, and economic sciences have all shown interest in studying ET FILIÈRES SYSTÈMES D’ÉLEVAGE Brazil, Uruguay the social group called family livestock farmers. The main characteristic of this ■ group, which is present in the Pampa biome in Southern Brazil and Uruguay, Accepted: 21 December 2014; Published: is beef cattle production based on family work on small lands, expressing an 25 March 2016 autonomous way of life which is, however, highly dependent on strong relations with the physical environment and marked by risk aversion. In this study we made a comparative analysis of vulnerability factors of family livestock farm- ing in Brazil and Uruguay. We also compared these social actors’ perceptions of risks, and the strategies built to mitigate threats. A survey was thus carried out and included 16 family livestock farmers’ interviews, eight in each country, near the cities of Santana do Livramento (Brazil) and Rivera (Uruguay). Although these cities are next to each other on each side of the border and thus present environmental similarities, we chose them because family farming was not sub- jected to the same political and economic conditions which might (or might not) have influenced farmers’ perceptions and reactions. Results showed that live- stock farmers were mainly affected by vulnerabilities arising from external ele- ments such as the climate (e.g. -

American Community Survey and Puerto Rico Community Survey
American Community Survey and Puerto Rico Community Survey 2016 Code List 1 TABLE OF CONTENTS ANCESTRY CODE LIST 3 FIELD OF DEGREE CODE LIST 25 GROUP QUARTERS CODE LIST 31 HISPANIC ORIGIN CODE LIST 32 INDUSTRY CODE LIST 35 LANGUAGE CODE LIST 44 OCCUPATION CODE LIST 80 PLACE OF BIRTH, MIGRATION, & PLACE OF WORK CODE LIST 95 RACE CODE LIST 105 2 Ancestry Code List ANCESTRY CODE WESTERN EUROPE (EXCEPT SPAIN) 001-099 . ALSATIAN 001 . ANDORRAN 002 . AUSTRIAN 003 . TIROL 004 . BASQUE 005 . FRENCH BASQUE 006 . SPANISH BASQUE 007 . BELGIAN 008 . FLEMISH 009 . WALLOON 010 . BRITISH 011 . BRITISH ISLES 012 . CHANNEL ISLANDER 013 . GIBRALTARIAN 014 . CORNISH 015 . CORSICAN 016 . CYPRIOT 017 . GREEK CYPRIOTE 018 . TURKISH CYPRIOTE 019 . DANISH 020 . DUTCH 021 . ENGLISH 022 . FAROE ISLANDER 023 . FINNISH 024 . KARELIAN 025 . FRENCH 026 . LORRAINIAN 027 . BRETON 028 . FRISIAN 029 . FRIULIAN 030 . LADIN 031 . GERMAN 032 . BAVARIAN 033 . BERLINER 034 3 ANCESTRY CODE WESTERN EUROPE (EXCEPT SPAIN) (continued) . HAMBURGER 035 . HANNOVER 036 . HESSIAN 037 . LUBECKER 038 . POMERANIAN 039 . PRUSSIAN 040 . SAXON 041 . SUDETENLANDER 042 . WESTPHALIAN 043 . EAST GERMAN 044 . WEST GERMAN 045 . GREEK 046 . CRETAN 047 . CYCLADIC ISLANDER 048 . ICELANDER 049 . IRISH 050 . ITALIAN 051 . TRIESTE 052 . ABRUZZI 053 . APULIAN 054 . BASILICATA 055 . CALABRIAN 056 . AMALFIAN 057 . EMILIA ROMAGNA 058 . ROMAN 059 . LIGURIAN 060 . LOMBARDIAN 061 . MARCHE 062 . MOLISE 063 . NEAPOLITAN 064 . PIEDMONTESE 065 . PUGLIA 066 . SARDINIAN 067 . SICILIAN 068 . TUSCAN 069 4 ANCESTRY CODE WESTERN EUROPE (EXCEPT SPAIN) (continued) . TRENTINO 070 . UMBRIAN 071 . VALLE DAOSTA 072 . VENETIAN 073 . SAN MARINO 074 . LAPP 075 . LIECHTENSTEINER 076 . LUXEMBURGER 077 . MALTESE 078 . MANX 079 . -

Livestock Farming Embedded in Local Development: Functional Perspective En Recherche Agronomique Pour Le Développement to Alleviate Vulnerability of Rural Communities
Contents Sommaire LiFLoD Livestock Farming & Local Development Rosario, Santa Fe, Argentina 31 March – 1 April 2011 www.liflod.org Workshop held during the 9th International Rangeland Congress ISSN 1951-6711 Publication du Centre de coopération internationale 51-53 Livestock farming embedded in local development: Functional perspective en recherche agronomique pour le développement to alleviate vulnerability of rural communities. Tourrand J.-F., Waquil P.D., Sraïri M.T., http://revues.cirad.fr/index.php/REMVT http://www.cirad.fr/ Hubert B. (in English) Directeur de la publication / Publication Director: LIVESTOCK FARMING SYSTEMS AND VALUE CHAINS Michel Eddi, PDG / President & CEO SYSTÈMES D’ÉLEVAGE ET FILIÈRES Rédacteurs en chef / Editors-in-Chief: Gilles Balança, Denis Bastianelli, Frédéric Stachurski 55-59 Vulnerability of family livestock farming on the Livramento-Rivera border of Brazil and Uruguay: Comparative analysis. Vulnérabilité des éleveurs familiaux à la Rédacteurs associés / Associate Editors: Guillaume Duteurtre, Bernard Faye, Flavie Goutard, frontière entre Livramento et Rivera au Brésil et en Uruguay : analyse comparative. Waquil Vincent Porphyre P.D., Neske M.Z., Ribeiro C.M., Schlick F.E., Andreatta T., Perleberg C., Borba M.F.S., Trindade J.P., Carriquiry R., Malaquin I., Saravia A., Gonzales M., Claudino L.S.D. (in English) Coordinatrice d’édition / Publishing Coordinator: Marie-Cécile Maraval 61-67 Opportunism and persistence in milk production in the Brazilian Traductrices/Translators: Amazonia. Opportunisme et persistance dans la production de lait en Amazonie Marie-Cécile Maraval (anglais), brésilienne. De Carvalho S.A., Poccard-Chapuis R., Tourrand J.-F. (in English) Suzanne Osorio-da Cruz (espagnol) Webmestre/Webmaster: Christian Sahut 69-74 Rangeland management in the Qilian mountains, Tibetan plateau, China. -
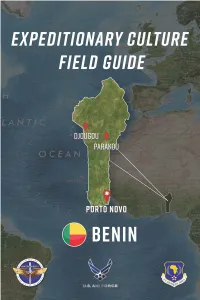
ECFG-Benin-Apr-19.Pdf
About this Guide This guide is designed to prepare you to deploy to culturally complex environments and achieve mission objectives. The fundamental information contained within will help you understand the decisive cultural dimension of your assigned location and gain skills necessary for success (Photo: Beninese medics perform routine physical examinations on local residents of Wanrarou, Benin). ECFG The guide consists of 2 parts: Part 1 introduces “Culture General,” the foundational knowledge you need to operate effectively in any global environment. Benin Part 2 presents “Culture Specific” Benin, focusing on unique cultural features of Beninese society and is designed to complement other pre-deployment training. It applies culture- general concepts to help increase your knowledge of your assigned deployment location (Photo: Beninese Army Soldier practices baton strikes during peacekeeping training with US Marine in Bembereke, Benin). For further information, visit the Air Force Culture and Language Center (AFCLC) website at www.airuniversity.af.edu/AFCLC/ or contact AFCLC’s Region Team at [email protected]. Disclaimer: All text is the property of the AFCLC and may not be modified by a change in title, content, or labeling. It may be reproduced in its current format with the expressed permission of the AFCLC. All photography is provided as a courtesy of the US government, Wikimedia, and other sources as indicated. GENERAL CULTURE PART 1 – CULTURE GENERAL What is Culture? Fundamental to all aspects of human existence, culture shapes the way humans view life and functions as a tool we use to adapt to our social and physical environments. A culture is the sum of all of the beliefs, values, behaviors, and symbols that have meaning for a society. -

Check List 4(4): 434–438, 2008. ISSN: 1809-127X
Check List 4(4): 434–438, 2008. ISSN: 1809-127X NOTES ON GEOGRAPHIC DISTRIBUTION Reptilia, Gekkonidae, Hemidactylus mabouia, Tarentola mauritanica: Distribution extension and anthropogenic dispersal Diego Baldo 1 Claudio Borteiro 2 Francisco Brusquetti 3 José Eduardo García 4 Carlos Prigioni 5 1 Universidad Nacional de Misiones, Facultad de Ciencias Exactas Químicas y Naturales. Félix de Azara 1552, (3300). Posadas, Misiones, Argentina. E-mail: [email protected] 2 Río de Janeiro 4058, 12800, Montevideo, Uruguay. 3 Instituto de Investigación Biológica del Paraguay (IIBP). Del Escudo 1607. 1429. Asunción, Paraguay. 4Dirección Nacional de Aduanas, La Coronilla, Rocha, Uruguay. Leopoldo Fernández s/n, La Coronilla, Rocha, Uruguay 5Museo Nacional de Historia Natural y Antropología. 25 de Mayo 582, Montevideo, Uruguay. The gekkonid genera Hemidactylus and Tarentola also found in an urban area (Achaval and Gudynas are composed by small sized lizards, noticeably 1983). able to perform long distance natural and anthropogenic dispersal, followed by colonization In this work we present new records of both of new areas (Kluge 1969, Vanzolini 1978, species in Uruguay, some of them associated to Carranza et al. 2000, Vences et al. 2004). Newly accidental anthropogenic dispersal, new records of introduced gecko species, at least of the genus H. mabouia in Argentina, and the first record of Hemidactylus, were reported as capable of the H. mabouia for Paraguay. Vouchers are displacing native ones (Hanley et al. 1998, Dame deposited at Colección Diego Baldo, housed at and Petren 2006, Rivas Fuenmayor et al. 2005). Museo de La Plata, Argentina (MLP DB), Colección Zoológica de la Facultad de Ciencias Interestingly, human related translocations aided Exactas y Naturales, Asunción, Paraguay some of these invasive lizards to have currently an (CZCEN), Museo Nacional de Historia Natural almost cosmopolitan distribution in tropical and de Montevideo (MNHN, currently Museo temperate regions (Vences et al.