Chloroplast and Nuclear Gene Sequences Indicatelate
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
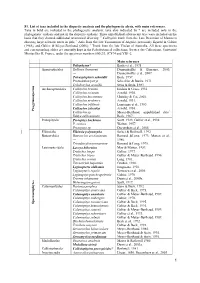
S1. List of Taxa Included in the Disparity Analysis and the Phylogenetic Alysis, with Main References
S1. List of taxa included in the disparity analysis and the phylogenetic alysis, with main references. Taxa in bold are included in the phylogenetic analysis; taxa also indicated by * are included only in the phylogenetic analysis and not in the disparity analysis. Three unpublished arborescent taxa were included on the basis that they showed additional anatomical diversity. 1 Callixylon trunk from the Late Devonian of Marrocco showing large sclerotic nests in pith; 2 Axis from the late Tournaisian of Algeria, previously figured in Galtier (1988), and Galtier & Meyer-Berthaud (2006); 3 Trunk from the late Viséan of Australia. All these specimens and corresponding slides are currently kept in the Paleobotanical collections, Service des Collections, Université Montpellier II, France, under the specimen numbers 600/2/3, JC874 and YB1-2. Main reference Psilophyton* Banks et al., 1975 Aneurophytales Rellimia thomsonii Dannenhoffer & Bonamo, 2003; --- Dannenhoffer et al., 2007. Tetraxylopteris schmidtii Beck, 1957. Proteokalon petryi Scheckler & Banks, 1971. Triloboxylon arnoldii Stein & Beck, 1983. s m Archaeopteridales Callixylon brownii Hoskin & Cross, 1951. r e Callixylon erianum Arnold, 1930. p s o Callixylon huronensis Chitaley & Cai, 2001. n Callixylon newberry Arnold, 1931. m y g Callixylon trifilievii Lemoigne et al., 1983. o r Callixylon zalesskyi Arnold, 1930. P Callixylon sp. Meyer-Berthaud, unpublished data1. Eddya sullivanensis Beck, 1967. Protopityales Protopitys buchiana Scott, 1923; Galtier et al., 1998. P. scotica Walton, 1957. Protopitys sp. Decombeix et al., 2005. Elkinsiales Elkinsia polymorpha Serbet & Rothwell, 1992. Buteoxylales Buteoxylon gordonianum Barnard &Long, 1973; Matten et al., --- 1980. Triradioxylon primaevum Barnard & Long, 1975. Lyginopteridales Laceya hibernica May & Matten, 1983. Tristichia longii Galtier, 1977. -

Identification of Conifer Trees in Iowa This Publication Is Designed to Help Identify the Most Common Trees Found in Iowa
Identification of Conifer Trees in Iowa This publication is designed to help identify the most common trees found in Iowa. It is based on vegetative characteristics including leaves, fruit, and bark. It is neither complete nor without possible oversights. Separate species are grouped by similar characteristics, mainly based on type and arrangement of leaves. These groups are; awl- or scale- like needles; single needles, flattened with rounded tips; single needles, square in cross section, with pointed tips; and needles in bundles or fasticles of two or more. Remember, vegetative character- istics are quite variable; use more than one specimen for comparison. Awl- or scale-like needles Juniperus Virginiana Eastern Red Cedar Leaves are dark green; leaves are both awl- and scale-like; cone is dark blue and berry-like. Thuja occidentalis Northern White Cedar Leaves are flattened and only of the scale type; cones have 4-6 scales; foliage is light green. Juniperus communis Common Juniper Leaves are awl shaped; cone is dark blue and berry-like. Pm-1383 | May 1996 Single needles, flattened with rounded tips Pseudotsuga menziesii Douglas Fir Needles occur on raised pegs; 3/4-11/4 inches in length; cones have 3-pointed bracts between the cone scales. Abies balsamea Abies concolor Balsam Fir White (Concolor) Fir Needles are blunt and notched at Needles are somewhat pointed, the tip; 3/4-11/2 inches in length. curved towards the branch top and 11/2-3 inches in length; silver green in color. Single needles, Picea abies Norway Spruce square in cross Needles are 1/2-1 inch long; section, with needles are dark green; foliage appears to droop or weep; cone pointed tips is 4-7 inches long. -

The Effects of Sulphur Dioxide on Selected Hepatics" (1978)
Eastern Illinois University The Keep Masters Theses Student Theses & Publications 1978 The ffecE ts of Sulphur Dioxide on Selected Hepatics Steven L. Gatchel Eastern Illinois University This research is a product of the graduate program in Botany at Eastern Illinois University. Find out more about the program. Recommended Citation Gatchel, Steven L., "The Effects of Sulphur Dioxide on Selected Hepatics" (1978). Masters Theses. 3192. https://thekeep.eiu.edu/theses/3192 This is brought to you for free and open access by the Student Theses & Publications at The Keep. It has been accepted for inclusion in Masters Theses by an authorized administrator of The Keep. For more information, please contact [email protected]. PAPF-:R CERTIFICATE #2 TO: Graduate Degree Candidates who have written formal theses. SUBJECT: Permission to reproduce theses. The University Library is receiving a ' number of requests from other institutions asking permission to reproduce dissertations for inclusion in their library holdings. Although no copyright laws are involved, we feel that professional courtesy demands that permission be obtained from the author before we allow theses to be copied. Please sign one of the following statements: Booth Library of Eastern Illinois University has my permission to lend my thesis to a reputable college or university for the purpose of copying it for inclusion in that institution's library or research holdings. Date Author I respectfully request Booth Library of .Eastern Illinois University not allow my thesis be reproduced because---------------- Date Author pdm THE EFFECTS OF SULPHUR DIOXIDE ON SELECTED HEPATICS (TITLE} BY Steven L. Gatchel THESIS SUBMITTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF Master of Science IN THE GRADUATE SCHOOL, EASTERN ILLINOIS UNIVERSITY CHARLESTON, ILLINOIS 1978 I HEREBY RECOMMEND THIS THESIS BE ACCEPTED AS FULFILLING THIS PART OF THE GRADUATE DEGREE CITED ABOVE ol}-~ d2/, 19"/f DATE ADVISER I/ 'Ouue~2\ 1\l~ DATE ' ~RTMENT WHEAD THE EFFECTS OF SULPHUR DIOXIDE ON SELECTED HEPATICS BY STEVEN L. -

Aquatic and Wet Marchantiophyta, Order Metzgeriales: Aneuraceae
Glime, J. M. 2021. Aquatic and Wet Marchantiophyta, Order Metzgeriales: Aneuraceae. Chapt. 1-11. In: Glime, J. M. Bryophyte 1-11-1 Ecology. Volume 4. Habitat and Role. Ebook sponsored by Michigan Technological University and the International Association of Bryologists. Last updated 11 April 2021 and available at <http://digitalcommons.mtu.edu/bryophyte-ecology/>. CHAPTER 1-11: AQUATIC AND WET MARCHANTIOPHYTA, ORDER METZGERIALES: ANEURACEAE TABLE OF CONTENTS SUBCLASS METZGERIIDAE ........................................................................................................................................... 1-11-2 Order Metzgeriales............................................................................................................................................................... 1-11-2 Aneuraceae ................................................................................................................................................................... 1-11-2 Aneura .......................................................................................................................................................................... 1-11-2 Aneura maxima ............................................................................................................................................................ 1-11-2 Aneura mirabilis .......................................................................................................................................................... 1-11-7 Aneura pinguis .......................................................................................................................................................... -

Ecophysiology of Four Co-Occurring Lycophyte Species: an Investigation of Functional Convergence
Research Article Ecophysiology of four co-occurring lycophyte species: an investigation of functional convergence Jacqlynn Zier, Bryce Belanger, Genevieve Trahan and James E. Watkins* Department of Biology, Colgate University, Hamilton, NY 13346, USA Received: 22 June 2015; Accepted: 7 November 2015; Published: 24 November 2015 Associate Editor: Tim J. Brodribb Citation: Zier J, Belanger B, Trahan G, Watkins JE. 2015. Ecophysiology of four co-occurring lycophyte species: an investigation of functional convergence. AoB PLANTS 7: plv137; doi:10.1093/aobpla/plv137 Abstract. Lycophytes are the most early divergent extant lineage of vascular land plants. The group has a broad global distribution ranging from tundra to tropical forests and can make up an important component of temperate northeast US forests. We know very little about the in situ ecophysiology of this group and apparently no study has eval- uated if lycophytes conform to functional patterns expected by the leaf economics spectrum hypothesis. To determine factors influencing photosynthetic capacity (Amax), we analysed several physiological traits related to photosynthesis to include stomatal, nutrient, vascular traits, and patterns of biomass distribution in four coexisting temperate lycophyte species: Lycopodium clavatum, Spinulum annotinum, Diphasiastrum digitatum and Dendrolycopodium dendroi- deum. We found no difference in maximum photosynthetic rates across species, yet wide variation in other traits. We also found that Amax was not related to leaf nitrogen concentration and is more tied to stomatal conductance, suggestive of a fundamentally different sets of constraints on photosynthesis in these lycophyte taxa compared with ferns and seed plants. These findings complement the hydropassive model of stomatal control in lycophytes and may reflect canaliza- tion of function in this group. -

X. the Conifers and Ginkgo
X. The Conifers and Ginkgo Now we turn our attention to the Coniferales, another great assemblage of seed plants. First let's compare the conifers with the cycads: Cycads Conifers few apical meristems per plant many apical meristems per plant leaves pinnately divided leaves undivided wood manoxylic wood pycnoxylic seeds borne on megaphylls seeds borne on stems We should also remember that these two groups have a lot in common. To begin with, they are both groups of woody seed plants. They are united by a small set of derived features: 1) the basic structure of the stele (a eustele or a sympodium, two words for the same thing) and no leaf gaps 2) the design of the apical meristem (many initials, subtended by a slowly dividing group of cells called the central mother zone) 3) the design of the tracheids (circular-bordered pits with a torus) We have three new seed plant orders to examine this week: A. Cordaitales This is yet another plant group from the coal forest. (Find it on the Peabody mural!) The best-known genus, Cordaites, is a tree with pycnoxylic wood bearing leaves up to about a foot and a half long and four inches wide. In addition, these trees bore sporangia (micro- and mega-) in strobili in the axils of these big leaves. The megasporangia were enclosed in ovules. Look at fossils of leaves and pollen-bearing shoots of Cordaites. The large, many-veined megaphylls are ancestral to modern pine needles; the shoots are ancestral to pollen-bearing strobili of modern conifers. 67 B. -

Laurentian-Acadian Alkaline Conifer-Hardwood Swamp
Laurentian-Acadian Alkaline Conifer-Hardwood Swamp Macrogroup: Northern Swamp yourStateNatural Heritage Ecologist for more information about this habitat. This is modeledmap a distributiononbased current and is data nota substitute for field inventory. based Contact © Elizabeth Thompson (Vermont Land Trust) Description: A forested swamp of alkaline wetlands associated with limestone or other calcareous substrate in the northern part of the glaciated northeast. Northern white cedar is often present and may dominate the canopy or be mixed with other conifers or with deciduous trees, most commonly red maple or black ash. Some examples can be almost entirely deciduous and dominated by black ash. Red-osier dogwood is a common shrub. The herb layer tends to be more diverse than in acidic swamps, due to higher pH and nutrient level. Small open fenny areas may occur within the wetland. The moss layer is often extensive and diverse. Seepage may influence parts of the wetland, but the hydrology is State Distribution: CT, MA, ME, NH, NY, VT dominated by the basin setting. Total Habitat Acreage: 921,478 Ecological Setting and Natural Processes: Percent Conserved: 19.5% These forested wetlands are uncommon in the glaciated State State GAP 1&2 GAP 3 Unsecured northeast except in areas with extensive limestone or similar State Habitat % Acreage (acres) (acres) (acres) substrate. The substrate is typically mineral soil, but there ME 56% 520,121 14,203 60,307 445,611 may be some peat, and there is often direct contact with NY 38% 345,750 49,536 44,764 251,450 alkaline groundwater. VT 5% 43,899 1,177 4,786 37,935 NH 1% 7,363 2,054 1,013 4,295 MA 0% 4,261 643 1,267 2,350 CT 0% 86 0 0 86 Similar Habitat Types: Similar to North-Central Interior and Appalachian Rich Swamp, but with a flora characteristic of a cooler climate. -

Ancient Noeggerathialean Reveals the Seed Plant Sister Group Diversified Alongside the Primary Seed Plant Radiation
Ancient noeggerathialean reveals the seed plant sister group diversified alongside the primary seed plant radiation Jun Wanga,b,c,1, Jason Hiltond,e, Hermann W. Pfefferkornf, Shijun Wangg, Yi Zhangh, Jiri Beki, Josef Pšenickaˇ j, Leyla J. Seyfullahk, and David Dilcherl,m,1 aState Key Laboratory of Palaeobiology and Stratigraphy, Nanjing Institute of Geology and Palaeontology, Chinese Academy of Sciences, Nanjing 210008, China; bCenter for Excellence in Life and Paleoenvironment, Chinese Academy of Sciences, Nanjing 210008, China; cUniversity of Chinese Academy of Sciences, Shijingshan District, Beijing 100049, China; dSchool of Geography, Earth and Environmental Sciences, University of Birmingham, Edgbaston, Birmingham B15 2TT, United Kingdom; eBirmingham Institute of Forest Research, University of Birmingham, Edgbaston, Birmingham B15 2TT, United Kingdom; fDepartment of Earth and Environmental Science, University of Pennsylvania, Philadelphia, PA 19104-6316; gState Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Xiangshan, Beijing 100093, China; hCollege of Paleontology, Shenyang Normal University, Key Laboratory for Evolution of Past Life in Northeast Asia, Ministry of Natural Resources, Shenyang 110034, China; iDepartment of Palaeobiology and Palaeoecology, Institute of Geology v.v.i., Academy of Sciences of the Czech Republic, 165 00 Praha 6, Czech Republic; jCentre of Palaeobiodiversity, West Bohemian Museum in Plzen, 301 36 Plzen, Czech Republic; kDepartment of Paleontology, Geozentrum, University of Vienna, 1090 Vienna, Austria; lIndiana Geological and Water Survey, Bloomington, IN 47404; and mDepartment of Geology and Atmospheric Science, Indiana University, Bloomington, IN 47405 Contributed by David Dilcher, September 10, 2020 (sent for review July 2, 2020; reviewed by Melanie Devore and Gregory J. -

California Partners in Flight the USDA Forest Service Klamath Bird Observatory and PRBO Conservation Science
The Coniferous Forest Bird Conservation Plan A Strategy for Protecting and Managing Coniferous Forest Habitats and Associated Birds in California Version 1.1 March 2002 A project of California Partners in Flight The USDA Forest Service Klamath Bird Observatory and PRBO Conservation Science Conservation Plan Lead Authors: John C. Robinson, USDA Forest Service John Alexander, Klamath Bird Observatory Conservation Plan Supporting Authors, PRBO Conservation Science: Sue Abbott Diana Humple Grant Ballard Melissa Pitkin Dan Barton Sandy Scoggin Gregg Elliott Diana Stralberg Sacha Heath Focal Species Account Authors: Black-backed Woodpecker – Kerry Farris Black-throated Gray Warbler – Tina Mark, USDA Forest Service Brown Creeper – Danielle LeFer, San Francisco Bay Bird Observatory Dark-eyed Junco – Jim DeStaebler, PRBO Conservation Science Flammulated Owl – Susan Yasuda, USDA Forest Service Fox Sparrow – Anne King, EDAW, Inc. Golden-crowned Kinglet – John C. Robinson, USDA Forest Service MacGillivray's Warbler – Chris Otahal, USDA Forest Service Olive-sided Flycatcher – Paul Brandy, Endangered Species Recovery Program Pileated Woodpecker – John C. Robinson, USDA Forest Service Red-breasted Nuthatch – Tina Mark and John C. Robinson, USDA Forest Service Vaux's Swift – John Sterling, Jones and Stokes Associates Western Tanager – Cory Davis, USDA Forest Service Financial Contributors: USDA Forest Service Packard Foundation National Fish and Wildlife Foundation PRBO Conservation Science Klamath Bird Observatory Acknowledgements: California Partners in Flight wishes to thank everyone who helped write, promote, and produce this document. Special thanks to Laurie Fenwood, Geoffrey Geupel, Aaron Holmes, Genny Wilson, Ryan Burnett, and Doug Wallace, and to Sophie Webb for her cover illustration. Recommended Citation: CalPIF (California Partners in Flight). 2002. -

Archaeopteris Is the Earliest Known Modern Tree
letters to nature seawater nitrate mapping systems for use in open ocean and coastal waters. Deep-Sea Res. I 43, 1763± zones as important sites for the subsequent development of lateral 1775 (1996). 26. Obata, H., Karatani, H., Matsui, M. & Nakayama, E. Fundamental studies for chemical speciation in organs; and wood anatomy strategies that minimize the mechani- seawater with an improved analytical method. Mar. Chem. 56, 97±106 (1997). cal stresses caused by perennial branch growth. Acknowledgements. We thank G. Elrod, E. Guenther, C. Hunter, J. Nowicki and S. Tanner for the iron Archaeopteris is thought to have been an excurrent tree, with a analyses and assistance with sampling, and the crews of the research vessels Western Flyer and New Horizon single trunk producing helically arranged deciduous branches for providing valuable assistance at sea. Funding was provided by the David and Lucile Packard 7 Foundation through MBARI and by the National Science Foundation. growing almost horizontally . All studies relating to the develop- ment of Archaeopteris support the view that these ephemeral Correspondence and requests for materials should be addressed to K.S.J. (e-mail: johnson@mlml. calstate.edu). branches arise from the pseudomonopodial division of the trunk apex8±11. Apical branching also characterizes all other contempora- neous non-seed-plant taxa including those that had also evolved an arborescent habit, such as lepidosigillarioid lycopsids and cladoxy- Archaeopteris is the earliest lalean ferns. This pattern, which can disadvantage the tree if the trunk apex is damaged, contrasts with the axillary branching knownmoderntree reported in early seed plants12. Analysis of a 4 m-long trunk from the Famennian of Oklahoma has shown that Archaeopteris may Brigitte Meyer-Berthaud*, Stephen E. -

Classification, Molecular Phylogeny, Divergence Time, And
The JapaneseSocietyJapanese Society for Plant Systematics ISSN 1346-7565 Acta Phytotax. Geobot. 56 (2): 111-126 (2005) Invited article and Classification,MolecularPhylogeny,DivergenceTime, Morphological Evolution of Pteridophytes with Notes on Heterospory and and Monophyletic ParaphyleticGroups MASAHIRO KATO* Department ofBiotogicat Sciences,Graduate Schoot ofScience,Universitv. of7bkyo, Hongo, 7bk)]o IJ3- O033, lapan Pteridophytes are free-sporing vascular land plants that evolutionarily link bryophytes and seed plants. Conventiona], group (taxon)-based hierarchic classifications ofptcridophytes using phenetic characters are briefiy reviewcd. Review is also made for recent trcc-based cladistic analyses and molecular phy- logenetic analyses with increasingly large data sets ofmultiplc genes (compared to single genes in pre- vious studies) and increasingly large numbers of spccies representing major groups of pteridophytes (compared to particular groups in previous studies), and it is cxtended to most recent analyses of esti- mating divergcnce times ofpteridephytes, These c]assifications, phylogenetics, and divergcncc time esti- mates have improved our understanding of the diversity and historical structure of pteridophytes. Heterospory is noted with referencc to its origins, endospory, fertilization, and dispersal. Finally, menophylctic and paraphyletic groups rccently proposed or re-recognized are briefly dcscribcd. Key words: classification, divergence timc estimate. fems,heterospory, molecular phylogcny, pteri- dophytcs. Morphological -

Diversity and Evolution of the Megaphyll in Euphyllophytes
G Model PALEVO-665; No. of Pages 16 ARTICLE IN PRESS C. R. Palevol xxx (2012) xxx–xxx Contents lists available at SciVerse ScienceDirect Comptes Rendus Palevol w ww.sciencedirect.com General palaeontology, systematics and evolution (Palaeobotany) Diversity and evolution of the megaphyll in Euphyllophytes: Phylogenetic hypotheses and the problem of foliar organ definition Diversité et évolution de la mégaphylle chez les Euphyllophytes : hypothèses phylogénétiques et le problème de la définition de l’organe foliaire ∗ Adèle Corvez , Véronique Barriel , Jean-Yves Dubuisson UMR 7207 CNRS-MNHN-UPMC, centre de recherches en paléobiodiversité et paléoenvironnements, 57, rue Cuvier, CP 48, 75005 Paris, France a r t i c l e i n f o a b s t r a c t Article history: Recent paleobotanical studies suggest that megaphylls evolved several times in land plant st Received 1 February 2012 evolution, implying that behind the single word “megaphyll” are hidden very differ- Accepted after revision 23 May 2012 ent notions and concepts. We therefore review current knowledge about diverse foliar Available online xxx organs and related characters observed in fossil and living plants, using one phylogenetic hypothesis to infer their origins and evolution. Four foliar organs and one lateral axis are Presented by Philippe Taquet described in detail and differ by the different combination of four main characters: lateral organ symmetry, abdaxity, planation and webbing. Phylogenetic analyses show that the Keywords: “true” megaphyll appeared at least twice in Euphyllophytes, and that the history of the Euphyllophytes Megaphyll four main characters is different in each case. The current definition of the megaphyll is questioned; we propose a clear and accurate terminology in order to remove ambiguities Bilateral symmetry Abdaxity of the current vocabulary.