Design and Development of Naïve Bayes Classifier
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Practical Statistics for Particle Physics Lecture 1 AEPS2018, Quy Nhon, Vietnam
Practical Statistics for Particle Physics Lecture 1 AEPS2018, Quy Nhon, Vietnam Roger Barlow The University of Huddersfield August 2018 Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 1 / 34 Lecture 1: The Basics 1 Probability What is it? Frequentist Probability Conditional Probability and Bayes' Theorem Bayesian Probability 2 Probability distributions and their properties Expectation Values Binomial, Poisson and Gaussian 3 Hypothesis testing Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 2 / 34 Question: What is Probability? Typical exam question Q1 Explain what is meant by the Probability PA of an event A [1] Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 3 / 34 Four possible answers PA is number obeying certain mathematical rules. PA is a property of A that determines how often A happens For N trials in which A occurs NA times, PA is the limit of NA=N for large N PA is my belief that A will happen, measurable by seeing what odds I will accept in a bet. Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 4 / 34 Mathematical Kolmogorov Axioms: For all A ⊂ S PA ≥ 0 PS = 1 P(A[B) = PA + PB if A \ B = ϕ and A; B ⊂ S From these simple axioms a complete and complicated structure can be − ≤ erected. E.g. show PA = 1 PA, and show PA 1.... But!!! This says nothing about what PA actually means. Kolmogorov had frequentist probability in mind, but these axioms apply to any definition. Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 5 / 34 Classical or Real probability Evolved during the 18th-19th century Developed (Pascal, Laplace and others) to serve the gambling industry. -

Linear Models for Classification
CS480 Introduction to Machine Learning Linear Models for Classification Edith Law Based on slides from Joelle Pineau Probabilistic Framework for Classification As we saw in the last lecture, learning can be posed a problem of statistical inference. The premise is that for any learning problem, if we have access to the underlying probability distribution D, then we could form a Bayes optimal classifier as: f (BO)(x)̂ = arg max D(x,̂ y)̂ y∈̂ Y But since we don't have access to D, we can instead estimate this distribution from data. The probabilistic framework allows you to choose a model to represent your data, and develop algorithms that have inductive biases that are closer to what you, as a designer, believe. 2 Classification by Density Estimation The most direct way to construct such a probability distribution is to select a family of parametric distributions. For example, we can select a Gaussian (or Normal) distribution. Gaussian is parametric: its parameters are its mean and variance. The job of learning is to infer which parameters are “best” in terms of describing the observed training data. In density estimation, we assume that the sample data is drawn independently from the same distribution D. That is, your nth sample (xn, yn) from D does not depend on the previous n-1 samples. This is called the i.i.d. (independently and identically distributed) assumption. 3 Consider the Problem of Flipping a Biased Coin You are flipping a coin, and you want to find out if it is biased. This coin has some unknown probability of β of coming up Head (H), and some probability ( 1 − β ) of coming up Tail (T). -
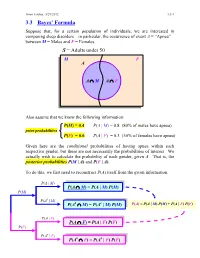
3.3 Bayes' Formula
Ismor Fischer, 5/29/2012 3.3-1 3.3 Bayes’ Formula Suppose that, for a certain population of individuals, we are interested in comparing sleep disorders – in particular, the occurrence of event A = “Apnea” – between M = Males and F = Females. S = Adults under 50 M F A A ∩ M A ∩ F Also assume that we know the following information: P(M) = 0.4 P(A | M) = 0.8 (80% of males have apnea) prior probabilities P(F) = 0.6 P(A | F) = 0.3 (30% of females have apnea) Given here are the conditional probabilities of having apnea within each respective gender, but these are not necessarily the probabilities of interest. We actually wish to calculate the probability of each gender, given A. That is, the posterior probabilities P(M | A) and P(F | A). To do this, we first need to reconstruct P(A) itself from the given information. P(A | M) P(A ∩ M) = P(A | M) P(M) P(M) P(Ac | M) c c P(A ∩ M) = P(A | M) P(M) P(A) = P(A | M) P(M) + P(A | F) P(F) P(A | F) P(A ∩ F) = P(A | F) P(F) P(F) P(Ac | F) c c P(A ∩ F) = P(A | F) P(F) Ismor Fischer, 5/29/2012 3.3-2 So, given A… P(M ∩ A) P(A | M) P(M) P(M | A) = P(A) = P(A | M) P(M) + P(A | F) P(F) (0.8)(0.4) 0.32 = (0.8)(0.4) + (0.3)(0.6) = 0.50 = 0.64 and posterior P(F ∩ A) P(A | F) P(F) P(F | A) = = probabilities P(A) P(A | M) P(M) + P(A | F) P(F) (0.3)(0.6) 0.18 = (0.8)(0.4) + (0.3)(0.6) = 0.50 = 0.36 S Thus, the additional information that a M F randomly selected individual has apnea (an A event with probability 50% – why?) increases the likelihood of being male from a prior probability of 40% to a posterior probability 0.32 0.18 of 64%, and likewise, decreases the likelihood of being female from a prior probability of 60% to a posterior probability of 36%. -

On Discriminative Bayesian Network Classifiers and Logistic Regression
On Discriminative Bayesian Network Classifiers and Logistic Regression Teemu Roos and Hannes Wettig Complex Systems Computation Group, Helsinki Institute for Information Technology, P.O. Box 9800, FI-02015 HUT, Finland Peter Gr¨unwald Centrum voor Wiskunde en Informatica, P.O. Box 94079, NL-1090 GB Amsterdam, The Netherlands Petri Myllym¨aki and Henry Tirri Complex Systems Computation Group, Helsinki Institute for Information Technology, P.O. Box 9800, FI-02015 HUT, Finland Abstract. Discriminative learning of the parameters in the naive Bayes model is known to be equivalent to a logistic regression problem. Here we show that the same fact holds for much more general Bayesian network models, as long as the corresponding network structure satisfies a certain graph-theoretic property. The property holds for naive Bayes but also for more complex structures such as tree-augmented naive Bayes (TAN) as well as for mixed diagnostic-discriminative structures. Our results imply that for networks satisfying our property, the condi- tional likelihood cannot have local maxima so that the global maximum can be found by simple local optimization methods. We also show that if this property does not hold, then in general the conditional likelihood can have local, non-global maxima. We illustrate our theoretical results by empirical experiments with local optimization in a conditional naive Bayes model. Furthermore, we provide a heuristic strategy for pruning the number of parameters and relevant features in such models. For many data sets, we obtain good results with heavily pruned submodels containing many fewer parameters than the original naive Bayes model. Keywords: Bayesian classifiers, Bayesian networks, discriminative learning, logistic regression 1. -

Naïve Bayes Classifier
CS 60050 Machine Learning Naïve Bayes Classifier Some slides taken from course materials of Tan, Steinbach, Kumar Bayes Classifier ● A probabilistic framework for solving classification problems ● Approach for modeling probabilistic relationships between the attribute set and the class variable – May not be possible to certainly predict class label of a test record even if it has identical attributes to some training records – Reason: noisy data or presence of certain factors that are not included in the analysis Probability Basics ● P(A = a, C = c): joint probability that random variables A and C will take values a and c respectively ● P(A = a | C = c): conditional probability that A will take the value a, given that C has taken value c P(A,C) P(C | A) = P(A) P(A,C) P(A | C) = P(C) Bayes Theorem ● Bayes theorem: P(A | C)P(C) P(C | A) = P(A) ● P(C) known as the prior probability ● P(C | A) known as the posterior probability Example of Bayes Theorem ● Given: – A doctor knows that meningitis causes stiff neck 50% of the time – Prior probability of any patient having meningitis is 1/50,000 – Prior probability of any patient having stiff neck is 1/20 ● If a patient has stiff neck, what’s the probability he/she has meningitis? P(S | M )P(M ) 0.5×1/50000 P(M | S) = = = 0.0002 P(S) 1/ 20 Bayesian Classifiers ● Consider each attribute and class label as random variables ● Given a record with attributes (A1, A2,…,An) – Goal is to predict class C – Specifically, we want to find the value of C that maximizes P(C| A1, A2,…,An ) ● Can we estimate -

Lecture 5: Naive Bayes Classifier 1 Introduction
CSE517A Machine Learning Spring 2020 Lecture 5: Naive Bayes Classifier Instructor: Marion Neumann Scribe: Jingyu Xin Reading: fcml 5.2.1 (Bayes Classifier, Naive Bayes, Classifying Text, and Smoothing) Application Sentiment analysis aims at discovering people's opinions, emo- tions, feelings about a subject matter, product, or service from text. Take some time to answer the following warm-up questions: (1) Is this a regression or classification problem? (2) What are the features and how would you represent them? (3) What is the prediction task? (4) How well do you think a linear model will perform? 1 Introduction Thought: Can we model p(y j x) without model assumptions, e.g. Gaussian/Bernoulli distribution etc.? Idea: Estimate p(y j x) from the data directly, then use the Bayes classifier. Let y be discrete, Pn i=1 I(xi = x \ yi = y) p(y j x) = Pn (1) i=1 I(xi = x) where the numerator counts all examples with input x that have label y and the denominator counts all examples with input x. Figure 1: Illustration of estimatingp ^(y j x) From Figure ??, it is clear that, using the MLE method, we can estimatep ^(y j x) as jCj p^(y j x) = (2) jBj But there is a big problem with this method: The MLE estimate is only good if there are many training vectors with the same identical features as x. 1 2 This never happens for high-dimensional or continuous feature spaces. Solution: Bayes Rule. p(x j y) p(y) p(y j x) = (3) p(x) Let's estimate p(x j y) and p(y) instead! 2 Naive Bayes We have a discrete label space C that can either be binary f+1; −1g or multi-class f1; :::; Kg. -

Numerical Physics with Probabilities: the Monte Carlo Method and Bayesian Statistics Part I for Assignment 2
Numerical Physics with Probabilities: The Monte Carlo Method and Bayesian Statistics Part I for Assignment 2 Department of Physics, University of Surrey module: Energy, Entropy and Numerical Physics (PHY2063) 1 Numerical Physics part of Energy, Entropy and Numerical Physics This numerical physics course is part of the second-year Energy, Entropy and Numerical Physics mod- ule. It is online at the EENP module on SurreyLearn. See there for assignments, deadlines etc. The course is about numerically solving ODEs (ordinary differential equations) and PDEs (partial differential equations), and introducing the (large) part of numerical physics where probabilities are used. This assignment is on numerical physics of probabilities, and looks at the Monte Carlo (MC) method, and at the Bayesian statistics approach to data analysis. It covers MC and Bayesian statistics, in that order. MC is a widely used numerical technique, it is used, amongst other things, for modelling many random processes. MC is used in fields from statistical physics, to nuclear and particle physics. Bayesian statistics is a powerful data analysis method, and is used everywhere from particle physics to spam-email filters. Data analysis is fundamental to science. For example, analysis of the data from the Large Hadron Collider was required to extract a most probable value for the mass of the Higgs boson, together with an estimate of the region of masses where the scientists think the mass is. This region is typically expressed as a range of mass values where the they think the true mass lies with high (e.g., 95%) probability. Many of you will be analysing data (physics data, commercial data, etc) for your PTY or RY, or future careers1 . -

The Bayesian Approach to Statistics
THE BAYESIAN APPROACH TO STATISTICS ANTHONY O’HAGAN INTRODUCTION the true nature of scientific reasoning. The fi- nal section addresses various features of modern By far the most widely taught and used statisti- Bayesian methods that provide some explanation for the rapid increase in their adoption since the cal methods in practice are those of the frequen- 1980s. tist school. The ideas of frequentist inference, as set out in Chapter 5 of this book, rest on the frequency definition of probability (Chapter 2), BAYESIAN INFERENCE and were developed in the first half of the 20th century. This chapter concerns a radically differ- We first present the basic procedures of Bayesian ent approach to statistics, the Bayesian approach, inference. which depends instead on the subjective defini- tion of probability (Chapter 3). In some respects, Bayesian methods are older than frequentist ones, Bayes’s Theorem and the Nature of Learning having been the basis of very early statistical rea- Bayesian inference is a process of learning soning as far back as the 18th century. Bayesian from data. To give substance to this statement, statistics as it is now understood, however, dates we need to identify who is doing the learning and back to the 1950s, with subsequent development what they are learning about. in the second half of the 20th century. Over that time, the Bayesian approach has steadily gained Terms and Notation ground, and is now recognized as a legitimate al- ternative to the frequentist approach. The person doing the learning is an individual This chapter is organized into three sections. -

Hybrid of Naive Bayes and Gaussian Naive Bayes for Classification: a Map Reduce Approach
International Journal of Innovative Technology and Exploring Engineering (IJITEE) ISSN: 2278-3075, Volume-8, Issue-6S3, April 2019 Hybrid of Naive Bayes and Gaussian Naive Bayes for Classification: A Map Reduce Approach Shikha Agarwal, Balmukumd Jha, Tisu Kumar, Manish Kumar, Prabhat Ranjan Abstract: Naive Bayes classifier is well known machine model could be used without accepting Bayesian probability learning algorithm which has shown virtues in many fields. In or using any Bayesian methods. Despite their naive design this work big data analysis platforms like Hadoop distributed and simple assumptions, Naive Bayes classifiers have computing and map reduce programming is used with Naive worked quite well in many complex situations. An analysis Bayes and Gaussian Naive Bayes for classification. Naive Bayes is manily popular for classification of discrete data sets while of the Bayesian classification problem showed that there are Gaussian is used to classify data that has continuous attributes. sound theoretical reasons behind the apparently implausible Experimental results show that Hybrid of Naive Bayes and efficacy of types of classifiers[5]. A comprehensive Gaussian Naive Bayes MapReduce model shows the better comparison with other classification algorithms in 2006 performance in terms of classification accuracy on adult data set showed that Bayes classification is outperformed by other which has many continuous attributes. approaches, such as boosted trees or random forests [6]. An Index Terms: Naive Bayes, Gaussian Naive Bayes, Map Reduce, Classification. advantage of Naive Bayes is that it only requires a small number of training data to estimate the parameters necessary I. INTRODUCTION for classification. The fundamental property of Naive Bayes is that it works on discrete value. -

LINEAR REGRESSION MODELS Likelihood and Reference Bayesian
{ LINEAR REGRESSION MODELS { Likeliho o d and Reference Bayesian Analysis Mike West May 3, 1999 1 Straight Line Regression Mo dels We b egin with the simple straight line regression mo del y = + x + i i i where the design p oints x are xed in advance, and the measurement/sampling i 2 errors are indep endent and normally distributed, N 0; for each i = i i 1;::: ;n: In this context, wehavelooked at general mo delling questions, data and the tting of least squares estimates of and : Now we turn to more formal likeliho o d and Bayesian inference. 1.1 Likeliho o d and MLEs The formal parametric inference problem is a multi-parameter problem: we 2 require inferences on the three parameters ; ; : The likeliho o d function has a simple enough form, as wenow show. Throughout, we do not indicate the design p oints in conditioning statements, though they are implicitly conditioned up on. Write Y = fy ;::: ;y g and X = fx ;::: ;x g: Given X and the mo del 1 n 1 n parameters, each y is the corresp onding zero-mean normal random quantity i plus the term + x ; so that y is normal with this term as its mean and i i i 2 variance : Also, since the are indep endent, so are the y : Thus i i 2 2 y j ; ; N + x ; i i with conditional density function 2 2 2 2 1=2 py j ; ; = expfy x =2 g=2 i i i for each i: Also, by indep endence, the joint density function is n Y 2 2 : py j ; ; = pY j ; ; i i=1 1 Given the observed resp onse values Y this provides the likeliho o d function for the three parameters: the joint density ab ove evaluated at the observed and 2 hence now xed values of Y; now viewed as a function of ; ; as they vary across p ossible parameter values. -

Paradoxes and Priors in Bayesian Regression
Paradoxes and Priors in Bayesian Regression Dissertation Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Agniva Som, B. Stat., M. Stat. Graduate Program in Statistics The Ohio State University 2014 Dissertation Committee: Dr. Christopher M. Hans, Advisor Dr. Steven N. MacEachern, Co-advisor Dr. Mario Peruggia c Copyright by Agniva Som 2014 Abstract The linear model has been by far the most popular and most attractive choice of a statistical model over the past century, ubiquitous in both frequentist and Bayesian literature. The basic model has been gradually improved over the years to deal with stronger features in the data like multicollinearity, non-linear or functional data pat- terns, violation of underlying model assumptions etc. One valuable direction pursued in the enrichment of the linear model is the use of Bayesian methods, which blend information from the data likelihood and suitable prior distributions placed on the unknown model parameters to carry out inference. This dissertation studies the modeling implications of many common prior distri- butions in linear regression, including the popular g prior and its recent ameliorations. Formalization of desirable characteristics for model comparison and parameter esti- mation has led to the growth of appropriate mixtures of g priors that conform to the seven standard model selection criteria laid out by Bayarri et al. (2012). The existence of some of these properties (or lack thereof) is demonstrated by examining the behavior of the prior under suitable limits on the likelihood or on the prior itself. -

Confidence Intervals for Probabilistic Network Classifiers
Computational Statistics & Data Analysis 49 (2005) 998–1019 www.elsevier.com/locate/csda Confidence intervals for probabilistic network classifiersଁ M. Egmont-Petersena,∗, A. Feeldersa, B. Baesensb aUtrecht University, Institute of Information and Computing Sciences, P. O. Box 80.089, 3508, TB Utrecht, The Netherlands bUniversity of Southampton, School of Management, UK Received 12 July 2003; received in revised form 28 June 2004 Available online 25 July 2004 Abstract Probabilistic networks (Bayesian networks) are suited as statistical pattern classifiers when the feature variables are discrete. It is argued that their white-box character makes them transparent, a requirement in various applications such as, e.g., credit scoring. In addition, the exact error rate of a probabilistic network classifier can be computed without a dataset. First, the exact error rate for probabilistic network classifiers is specified. Secondly, the exact sampling distribution for the conditional probability estimates in a probabilistic network classifier is derived. Each conditional probability is distributed according to the bivariate binomial distribution. Subsequently, an approach for computing the sampling distribution and hence confidence intervals for the posterior probability in a probabilistic network classifier is derived. Our approach results in parametric bootstrap confidence intervals. Experiments with general probabilistic network classifiers, the Naive Bayes classifier and tree augmented Naive Bayes classifiers (TANs) show that our approximation performs well. Also simulations performed with the Alarm network show good results for large training sets. The amount of computation required is exponential in the number of feature variables. For medium and large-scale classification problems, our approach is well suited for quick simulations.A running example from the domain of credit scoring illustrates how to actually compute the sampling distribution of the posterior probability.