Runa, F., Park, M. S., and Pryor, B. M. 2009. Ulocladium Systematics
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Nature and Effect of Alternaria Spp. Complex from Wheat Grain on Germination and Disease Transmission
Pak. J. Bot., 45(5): 1817-1824, 2013. NATURE AND EFFECT OF ALTERNARIA SPP. COMPLEX FROM WHEAT GRAIN ON GERMINATION AND DISEASE TRANSMISSION ANALÍA E. PERELLÓ1,2* AND SILVINA LARRÁN1 1CIDEFI (Centro de Investigaciones de Fitopatología) y Cátedra de Fitopatología 2CONICET-Facultad de Ciencias Agrarias y Forestales de la Universidad Nacional de La Plata, Calle 60 y 119 (1900) La Plata, Buenos Aires, Argentina. *Corresponding author’s e-mail: anaperello2@ yahoo.com.ar Abstract Diseases caused by Alternaria sp. are among the most common diseases of crops throughout the world. Alternaria sp. is a common component of the flora of wheat seed. Although isolation of Alternaria sp. from wheat (Triticum aestivum) seed has been reported in Argentina, development of the Alternaria blight in plants from infected seeds has not been demonstrated experimentally. Seed transmission of strains belonging to Alternaria tenuissima, A. alternata, A. infectoria, A. triticina, A. chlamydospora and related genera like Embellisia and Ulocladium sp. on wheat were investigated in the Argentinean growing area, on wheat cultivars Klein Escorpión and Buck Poncho. A. tenuissima was the dominant fungus in black pointed kernels. Transmission of all 42 seed-borne members of Alternaria complex from seeds to seedlings artificially inoculated was detected by trays seedling symptoms test. Among the fungi tested most isolates of Alternaria, Embellisia sp. and Ulocladium sp. produced distinct seed rot and seedling infection symptoms. This confirmed the seed-borne nature of these fungi. In each wheat cultivar tested inoculated seeds appreciably reduced their germination. The emerging coleoptile is externally infected by hyphal growth from the infected pericarp. -

43-09-405 and 407 Final Combine
Pak. J. Bot ., 44(6): 2093-2102, 2012. A NEW SPECIES OF TIAROSPORELLA AZADARICHTA AND NEW FUNGAL RECORDS ON AZADIRACHTA INDICA FROM PAKISTAN SYED QAISER ABBAS 1, TEHREEMA IFTIKHAR 1* , MUBASHIR NIAZ 1, IFTIKHAR ALI 1, NABILA IFTIKHAR 1 AND ALIA ABBAS 2 1Department of Botany, Government College University, Alama Iqbal Road, Faisalabad, Pakistan 2Department of Botany, Federal Urdu University, Karachi, Pakistan *Corresponding author’s e-mail: [email protected] Abstract A new species of Tiarosporella azadarichta has been described on Azadirachta indica and some new fungal records viz ., Diplozythiella bambusina , Ulocladium chartarum , Cladosporium nigrellum, Cladosporium oxysporum. Didymostilbe coffeae , Muellerella pygmaea, Lasiodiplodia paraphysaria, Monochaetinula terminalae, Trimmatostroma sp., and Epidermophyton floccosum are reported on Azadirachta indica for the first time from Pakistan. Introduction reason a detailed survey of the plant for fungi was under taken in this study. Azadirachta indica (as Azadirachta Juss, Melia azadirachta ) belong to the family Meliaceae. Neem is its Materials and Methods common name. It is important due to its commercial and medicinal value. Azadirachta indica is also an important Samples of Azadirachta indica were collected from plant for its potential of anti-fungal, anti-bacterial and the different areas of District Faisalabad and Gojra. The insecticidal activity. Decline of trees due to fungi are different areas were G.C. University. Faisalabad, increasing tremendously in Pakistan especially in Punjab Agriculture University Faisalabad, Gutwala forest (park) and Sindh (Javed et al ., 2004; Khanzada et al ., 2011; Fateh Faisalabad, Tandlianwala and Gojra City. Methods and et al., 2011; Rheman et al., 2011; Farooq et al., 2011). materials are the same as described Abbas et al ., (2010). -

An Unusual Haemoid Fungi: Ulocladium, As a Cause Of
Volume 2 Number 2 (June 2010) 95-97 CASE Report An unusual phaeoid fungi: Ulocladium, as a cause of chronic allergic fungal sinusitis Kaur R, Wadhwa A1,Gulati A2, Agrawal AK2 1Department of Microbiology. 2Department of Otorhinolaryngology, Maulana Azad Medical College, New Delhi, India. Received: April 2010, Accepted: May 2010. ABSTRACT Allergic fungal sinusitis (AFS) has been recognized as an important cause of chronic sinusitis commonly caused by Aspergillus spp. and various dematiaceous fungi like Bipolaris, Alternaria, Curvalaria, and etc. Ulocladium botrytis is a non pathogenic environmental dematiaceous fungi, which has been recently described as a human pathogen. Ulocladium has never been associated with allergic fungal sinusitis but it was identified as an etiological agent of AFS in a 35 year old immunocompetent female patient presenting with chronic nasal obstruction of several months duration to our hospital. The patient underwent FESS and the excised polyps revealed Ulocladium as the causative fungal agent. Keywords: Ulocladium, Phaeoid Fungi, Chronic Sinusitis. INTRODUCTION CASE REPORT Chronic allergic sinusitis is a common condition A 35 year old female patient presented to the ENT responsible for the development of nasal polyps, out patient department of the Lok Nayak Hospital, described as abnormal lesions that emanate from Delhi, with chronic nasal obstruction, excessive any portion of the nasal mucosa or Para nasal sneezing, nasal discharge and frontal headache since sinuses. They are commonly located in the middle several months. Nasal obstruction was of gradual meatus and ethmoid sinus and are present in onset, non progressive, more on the left side than 1-4% of the population (1). -

Genetic Variability in the Pistachio Late Blight Fungus, Alternaria Alternata
Mycol. Res. 105 (3): 300–306 (March 2001). Printed in the United Kingdom. 300 Genetic variability in the pistachio late blight fungus, Alternaria alternata Mallikarjuna K. ARADHYA*, Helen M. CHAN and Dan E. PARFITT Department of Pomology, University of California, One Shields Avenue, Davis, CA 95616, USA. E-mail: aradhya!ucdavis.edu Received 15 December 1999; accepted 21 August 2000 Genetic variation in the pistachio late blight fungus, Alternaria alternata, was investigated by restriction fragment length polymorphism (RFLP) in the rDNA region. Southern hybridization of EcoRI, HindIII, and XbaI digested fungal DNA with a RNA probe derived from Alt1, an rDNA clone isolated from a genomic library of the Japanese pear pathotype of A. alternata, revealed 34 different rDNA haplotypes among 56 isolates collected from four central valley locations in California. Analysis of molecular variation revealed a significant amount of genetic diversity within populations (85n8%), with only marginal variation accounting for differentiation among populations (14n2%, ΦST l 0n142). All isolates examined were highly pathogenic. The identity of the four geographic populations sampled was not evident in both cluster and principal component analyses, probably indicating either the selectively neutral nature of rDNA variation or prevalence of widespread gene flow among populations combined with uniform host-selection. INTRODUCTION 1998). Biochemical and molecular markers have been used to demonstrate the existence of multiple strains and to assess The genus Alternaria contains diverse and ubiquitous species of infraspecific variation in A. alternata at the protein and DNA fungi, including aggressive and opportunistic plant pathogens level (Petrunak & Christ 1992, Adachi et al. 1993, Weir et al. -

Molecular Systematics of the Marine Dothideomycetes
available online at www.studiesinmycology.org StudieS in Mycology 64: 155–173. 2009. doi:10.3114/sim.2009.64.09 Molecular systematics of the marine Dothideomycetes S. Suetrong1, 2, C.L. Schoch3, J.W. Spatafora4, J. Kohlmeyer5, B. Volkmann-Kohlmeyer5, J. Sakayaroj2, S. Phongpaichit1, K. Tanaka6, K. Hirayama6 and E.B.G. Jones2* 1Department of Microbiology, Faculty of Science, Prince of Songkla University, Hat Yai, Songkhla, 90112, Thailand; 2Bioresources Technology Unit, National Center for Genetic Engineering and Biotechnology (BIOTEC), 113 Thailand Science Park, Paholyothin Road, Khlong 1, Khlong Luang, Pathum Thani, 12120, Thailand; 3National Center for Biothechnology Information, National Library of Medicine, National Institutes of Health, 45 Center Drive, MSC 6510, Bethesda, Maryland 20892-6510, U.S.A.; 4Department of Botany and Plant Pathology, Oregon State University, Corvallis, Oregon, 97331, U.S.A.; 5Institute of Marine Sciences, University of North Carolina at Chapel Hill, Morehead City, North Carolina 28557, U.S.A.; 6Faculty of Agriculture & Life Sciences, Hirosaki University, Bunkyo-cho 3, Hirosaki, Aomori 036-8561, Japan *Correspondence: E.B. Gareth Jones, [email protected] Abstract: Phylogenetic analyses of four nuclear genes, namely the large and small subunits of the nuclear ribosomal RNA, transcription elongation factor 1-alpha and the second largest RNA polymerase II subunit, established that the ecological group of marine bitunicate ascomycetes has representatives in the orders Capnodiales, Hysteriales, Jahnulales, Mytilinidiales, Patellariales and Pleosporales. Most of the fungi sequenced were intertidal mangrove taxa and belong to members of 12 families in the Pleosporales: Aigialaceae, Didymellaceae, Leptosphaeriaceae, Lenthitheciaceae, Lophiostomataceae, Massarinaceae, Montagnulaceae, Morosphaeriaceae, Phaeosphaeriaceae, Pleosporaceae, Testudinaceae and Trematosphaeriaceae. Two new families are described: Aigialaceae and Morosphaeriaceae, and three new genera proposed: Halomassarina, Morosphaeria and Rimora. -

Thesis FINAL PRINT
Preface This study was conducted at the Department of Chemistry, Biotechnology and Food Science (IKBM), Norwegian University of Life Sciences (UMB) during November 2009 to November 2010. My supervisors were Professor Dr Arne Tronsmo and PhD student Md. Hafizur Rahman, Department of IKBM, UMB. I express my sincere appreciation and gratitude to Professor Arne Tronsmo and Dr. Linda Gordon Hjeljord for their dynamic guidance throughout the period of the study, constant encouragement, constructive criticism and valuable suggestion during preparation of the thesis. I also want to thank Md. Hafizur Rahman for his guidance during my research work. Moreover I express my sincere gratitude to Grethe Kobro and Else Maria Aasen and all other staffs and workers at IKBM, UMB for their helpful co-operation to complete the research work in the laboratory. Last but not the least; I feel the heartiest indebtedness to Sabine and my family members for their patient inspirations, sacrifices and never ending encouragement. Ås, January 2011 Latifur Rahman Shovan i Abstract This thesis has been focused on methods to control diseases caused by Botrytis cinerea. B. cinerea causes grey mould disease of strawberry and chickpea, as well as many other plants. The fungal isolates used were isolated from chickpea leaf (Gazipur, Bangladesh) or obtained from the Norwegian culture collections of Bioforsk (Ås) and IKBM (UMB). Both morphological and molecular characterization helped to identify the fungal isolates as Botrytis cinerea (B. cinerea 101 and B. cinerea-BD), Trichoderma atroviride, T. asperellum Alternaria brassicicola, and Mucor piriformis. The identity of one fungal isolate, which was obtained from the culture collection of Bioforsk under the name Microdochium majus, could not be confirmed in this study. -
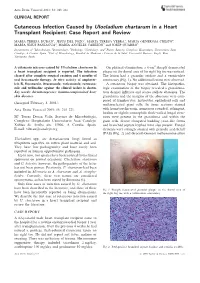
Cutaneous Infection Caused by Ulocladium Chartarum in a Heart Transplant Recipient: Case Report and Review
Acta Derm Venereol 2003; 83: 218–221 CLINICAL REPORT Cutaneous Infection Caused by Ulocladium chartarum in a Heart Transplant Recipient: Case Report and Review MARI´A TERESA DURA´ N1, JESU´ S DEL POZO2, MARI´A TERESA YEBRA3, MARI´A GENEROSA CRESPO4, MARI´A JESU´ S PANIAGUA4, MARI´A ANGELES CABEZO´ N5 and JOSEP GUARRO6 Departments of 1Microbiology, 2Dermatology, 3Pathology, 4Cardiology, and 5Plastic Surgery, Complexo Hospitalario Universitario Juan Canalejo, A Corun˜a, Spain, 6Unit of Microbiology, Facultat de Medicina i Cie`ncies de la Salut, Universitat Rovira i Virgili, Reus, Tarragona, Spain A cutaneous mycoses caused by Ulocladium chartarum in On physical examination, a 6-cm2 sharply demarcated a heart transplant recipient is reported. The infection plaque on the dorsal area of his right big toe was noticed. cleared after complete surgical excision and 6 months of The lesion had a granular surface and a vermiculate oral itraconazole therapy. In vitro activity of amphoter- consistency (Fig. 1). No additional lesions were observed. icin B, fluconazole, itraconazole, voriconazole, ravucona- A cutaneous biopsy was obtained. The histopatho- zole and terbinafine against the clinical isolate is shown. logic examination of the biopsy revealed a granuloma- Key words: dermatomycoses; immunocompromised host; tous dermal infiltrate and scarce stellate abscesses. The skin diseases. granuloma and the margins of the abscesses were com- posed of lymphocytes, histiocytes, epithelioid cells and (Accepted February 3, 2003.) multinucleated giant cells. In tissue sections stained Acta Derm Venereol 2003; 83: 218–221. with hematoxylin-eosin, numerous rounded, refringent, hyaline or slightly eosinophilic thick-walled fungal struc- Ma Teresa Dura´n Valle, Servicio de Microbiologı´a, tures were present in the granuloma and within the Complexo Hospitalario Universitario Juan Canalejo, giant cells. -

Identification and Characterization of Alternaria Species Causing Leaf Spot
Eur J Plant Pathol (2017) 149:401–413 DOI 10.1007/s10658-017-1190-0 Identification and characterization of Alternaria species causing leaf spot on cabbage, cauliflower, wild and cultivated rocket by using molecular and morphological features and mycotoxin production Ilenia Siciliano & Giovanna Gilardi & Giuseppe Ortu & Ulrich Gisi & Maria Lodovica Gullino & Angelo Garibaldi Accepted: 22 February 2017 /Published online: 11 March 2017 # Koninklijke Nederlandse Planteziektenkundige Vereniging 2017 Abstract Alternaria species are common pathogens of Keywords Crucifers . Toxins . Leaf spot . Tenuazonic fruit and vegetables able to produce secondary metabo- acid lites potentially affecting human health. Twenty-nine isolates obtained from cabbage, cauliflower, wild and cultivated rocket were characterized and identified Introduction based on sporulation pattern and virulence; the phylo- β genetic analysis was based on the -tubulin gene. Most Alternaria species are saprophytes and ubiquitous Isolates were identified as A. alternata, A. tenuissima, in the environment, however some are plant pathogenic, A. arborescens, A. brassicicola and A. japonica. inducing diseases on a large variety of economically Pathogenicity was evaluated on plants under greenhouse important crops like cereals, oil-crops, vegetables and conditions. Two isolates showed low level of virulence fruits (Pitt and Hocking, 1997). Most Alternaria spp. on cultivated rocket while the other isolates showed produce chains of conidia with transverse and longitu- medium or high level of virulence. Isolates were also dinal septa with a tapering apical cell. Conidial size, characterized for their mycotoxin production on a mod- presence and size of a beak, the pattern of catenation ified Czapek-Dox medium. Production of the five and longitudinal and transverse septation are key taxo- Alternaria toxins, tenuazonic acid, alternariol, nomic features for this genus (Joly 1964; Ellis 1971 and alternariol monomethyl ether, altenuene and tentoxin 1976, Simmons 1992). -

Novel Dematiaceous Hyphomycetes
STUDIES IN MYCOLOGY 50: 109–118. 2004. Novel dematiaceous hyphomycetes * Emory G. Simmons 717 Thornwood Road, Crawfordsville, Indiana 47933, U.S.A. *Correspondence: Emory G. Simmons, [email protected] Abstract: Novel taxa are described as Alternaria simsimi isolated from Sesamum indicum, distinguishing it from Alternaria sesami; Embellisia lolii from Lolium perenne; Nimbya perpunctulata from Alternanthera philoxeroides; Ulocladium arbores- cens from Pistacia vera; and Stemphylium symphyti from Symphytum × uplandicum. Taxonomic novelties: Alternaria simsimi E.G. Simmons sp. nov., Embellisia lolii E.G. Simmons & C.F. Hill sp. nov., Nimbya perpunctulata E.G. Simmons sp. nov., Stemphylium symphyti E.G. Simmons sp. nov., Ulocladium arborescens E.G. Simmons sp. nov. Key words: Alternaria, Embellisia, Nimbya, Ulocladium, Stemphylium, anamorphs, Sesamum, Lolium, Alternanthera, Pistacia, Symphytum. INTRODUCTION described and discussed on the basis of examination of colonies at 50× magnification and, at higher magnifi- It is a pleasure to be able to offer several new taxa in cations, of sporulation elements sampled at 5–7 d after honour of the centenary observances of the Centraal- inoculation. Each isolate or specimen discussed is bureau voor Schimmelcultures. These are dedicated to identified by a unique record/research number in the mycologist colleagues who were among my first format E.G.S. 00.000. associates during my earliest visits to CBS in Baarn, beginning in the late 1950s: Agathe van Beverwijk, Gerharda Bunschoten, G.A. de Vries, Amelia Stolk, TAXONOMIC PART Beatrice Schol-Schwarz, Maria Schipper, and Annie van der Plaats-Niterink. A novel taxon is presented for Alternaria: three species on sesame, two as patho- each of the five hyphomycetous genera that have been gens the major focus of my taxonomic work over the past Alternaria isolates from invaded tissues of Sesamum 50+ years. -

Monograph on Dematiaceous Fungi
Monograph On Dematiaceous fungi A guide for description of dematiaceous fungi fungi of medical importance, diseases caused by them, diagnosis and treatment By Mohamed Refai and Heidy Abo El-Yazid Department of Microbiology, Faculty of Veterinary Medicine, Cairo University 2014 1 Preface The first time I saw cultures of dematiaceous fungi was in the laboratory of Prof. Seeliger in Bonn, 1962, when I attended a practical course on moulds for one week. Then I handled myself several cultures of black fungi, as contaminants in Mycology Laboratory of Prof. Rieth, 1963-1964, in Hamburg. When I visited Prof. DE Varies in Baarn, 1963. I was fascinated by the tremendous number of moulds in the Centraalbureau voor Schimmelcultures, Baarn, Netherlands. On the other hand, I was proud, that El-Sheikh Mahgoub, a Colleague from Sundan, wrote an internationally well-known book on mycetoma. I have never seen cases of dematiaceous fungal infections in Egypt, therefore, I was very happy, when I saw the collection of mycetoma cases reported in Egypt by the eminent Egyptian Mycologist, Prof. Dr Mohamed Taha, Zagazig University. To all these prominent mycologists I dedicate this monograph. Prof. Dr. Mohamed Refai, 1.5.2014 Heinz Seeliger Heinz Rieth Gerard de Vries, El-Sheikh Mahgoub Mohamed Taha 2 Contents 1. Introduction 4 2. 30. The genus Rhinocladiella 83 2. Description of dematiaceous 6 2. 31. The genus Scedosporium 86 fungi 2. 1. The genus Alternaria 6 2. 32. The genus Scytalidium 89 2.2. The genus Aurobasidium 11 2.33. The genus Stachybotrys 91 2.3. The genus Bipolaris 16 2. -

PERSOONIAL R Eflections
Persoonia 23, 2009: 177–208 www.persoonia.org doi:10.3767/003158509X482951 PERSOONIAL R eflections Editorial: Celebrating 50 years of Fungal Biodiversity Research The year 2009 represents the 50th anniversary of Persoonia as the message that without fungi as basal link in the food chain, an international journal of mycology. Since 2008, Persoonia is there will be no biodiversity at all. a full-colour, Open Access journal, and from 2009 onwards, will May the Fungi be with you! also appear in PubMed, which we believe will give our authors even more exposure than that presently achieved via the two Editors-in-Chief: independent online websites, www.IngentaConnect.com, and Prof. dr PW Crous www.persoonia.org. The enclosed free poster depicts the 50 CBS Fungal Biodiversity Centre, Uppsalalaan 8, 3584 CT most beautiful fungi published throughout the year. We hope Utrecht, The Netherlands. that the poster acts as further encouragement for students and mycologists to describe and help protect our planet’s fungal Dr ME Noordeloos biodiversity. As 2010 is the international year of biodiversity, we National Herbarium of the Netherlands, Leiden University urge you to prominently display this poster, and help distribute branch, P.O. Box 9514, 2300 RA Leiden, The Netherlands. Book Reviews Mu«enko W, Majewski T, Ruszkiewicz- The Cryphonectriaceae include some Michalska M (eds). 2008. A preliminary of the most important tree pathogens checklist of micromycetes in Poland. in the world. Over the years I have Biodiversity of Poland, Vol. 9. Pp. personally helped collect populations 752; soft cover. Price 74 €. W. Szafer of some species in Africa and South Institute of Botany, Polish Academy America, and have witnessed the of Sciences, Lubicz, Kraków, Poland. -

Managing Alternaria Brown Spot by L.W
Managing Alternaria brown spot By L.W. Timmer, R. F. Reis, S.N. Mondal and N.A. Peres anker and greening may be problem for production of Fortune tan - the diseases that are on gerine in Spain. More recently, we Fig. 1. Alternaria brown spot lesions everyone’s minds, but we have confirmed the presence of the on a Minneola tangelo leaf. Note the still have to deal with the disease in Peru, where it is severe on characteristic necrosis running up Cmany everyday problems of produc - Minneolas, and in Iran, where it the veins. tion. Alternaria brown spot is the most causes problems mostly for Fortune for a few days on the grove floor, but serious disease of many popular tan - and Page production. then ceases as the leaves decay. Spores gerines and tangerine hybrids such as Tangerine cultivars, including Min - can also be produced on fruit and Minneola and Orlando tangelos, Mur - neolas, Orlandos, Novas, Lees, twigs, but are relatively few compared cotts, Sunburst, Novas and others. Ponkan, Murcotts, Dancy and many to the production on leaves. However, This disease probably requires more others, that are susceptible to ABS lesions on fruit and twigs may be im - intense management than any of the carry a gene for susceptibility to the portant in the overw-inter survival of other fungal diseases. toxin. That gene is dominant and all of the pathogen. Alternaria brown spot (ABS) is the offspring of susceptible parents are In the 2004-05 season, Alternaria caused by the fungus Alternaria alter - also susceptible. Many cultivars devel - pressure was very low and growers nata .