A Protein Microarray-Based in Vitro Transglutaminase Assay Platform For
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Ever 2018 Eupo 2018
European Association for Vision and Eye Research European University Professors of Ophthalmology EVER 2018 Annual Congress October 4-6, 2018 EUPO 2018 Course on Retina, Intraocular Inflammation & Uveitis October 3-4, 2018 Programme book Nice, France www.ever.be www.eupo.eu European Association for Vision and Eye Research EVER 20October 17-1919 in Nice, France www.ever.be 1 Table of contents Word from the president ....................................................................................................................................2 About EVER ..........................................................................................................................................................3 EVER Membership ...............................................................................................................................................4 Speakers’ affiliation to scientific sections .........................................................................................................5 Composition of the board 2018 .........................................................................................................................8 Venue ................................................................................................................................................................... 10 Congress information ....................................................................................................................................... 11 Programme information ....................................................................................................................................15 -

Short-Term Rapamycin Persistently Improves Cardiac Function After Cessation of Treatment in Aged Male and Female Mice
Short-term rapamycin persistently improves cardiac function after cessation of treatment in aged male and female mice. Ellen Quarles A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy University of Washington 2017 Reading Committee: Peter Rabinovitch, Chair Michael MacCoss David Marcinek Program Authorized to Offer Degree: Pathology © Copyright 2017 Ellen Quarles University of Washington Abstract Short-term rapamycin persistently improves cardiac function after cessation of treatment in aged male and female mice. Ellen Quarles Chair of the Supervisory Committee: Peter Rabinovitch, Professor and Vice Chair of Research Department of Pathology Cardiac aging is an intrinsic process that results in impaired cardiac function and dysregulation of cellular and molecular quality control mechanisms. These effects are evident in the decline of diastolic function, increase in left ventricular hypertrophy, metabolic substrate shifts, and alterations to the cardiac proteome. This thesis covers the quality control mechanisms that are associated with cardiac aging, results from an anti-aging intervention in aged mice, and a review of mitochondrial dysfunction in the heart. Chapter one is a review of the quality control mechanisms in aging myocardium. Chapter two consists of the results of several mouse experiments that compare the cardiac function, proteomes, and metabolomes of aged and young controls, along with rapamycin treated aged mice. The novelty of this study comes from the inclusion of a group of animals treated only transiently with the drug, then followed for eight weeks post-drug-removal. This persistence cohort may hold clues to deriving long-lasting benefits of rapamycin with only transient treatment. -

A Case of Long QT Syndrome
CASE REPORT A case of long QT syndrome: challenges on a bumpy road Peter Magnusson1,2 & Per-Erik Gustafsson2 1Cardiology Research Unit, Department of Medicine, Karolinska Institutet, Stockholm SE-171 76, Sweden 2Centre for Research and Development, Uppsala University/Region Gavleborg,€ Gavle€ SE-801 87, Sweden Correspondence Key Clinical Message Peter Magnusson, Cardiology Research Unit, Department of Medicine, Karolinska Beta-agonist treatment during pregnancy may unmask the diagnosis of long QT Institutet, Karolinska University Hospital/ syndrome. The QT prolongation can result in functional AV block. A history of Solna, SE-171 76 Stockholm, Sweden. Tel: seizure and/or sudden death in a family member should raise suspicion of ven- +46(0)705 089407; Fax: +46(0)26 154255; tricular tachycardia. More than one mutation may coexist. Refusal of beta- E-mail: [email protected] blocker therapy complicates risk stratification. Funding Information Keywords No sources of funding were declared for this Genetic, implantable cardioverter–defibrillator, long QT syndrome, pregnancy, study. premature ventricular complex, risk stratification, sudden cardiac death. Received: 16 December 2016; Revised: 29 March 2017; Accepted: 4 April 2017 Clinical Case Reports 2017; 5(6): 954–960 doi: 10.1002/ccr3.985 Introduction experienced palpitations, and her ECG was abnormal, revealing PVCs, atrioventricular (AV) 2:1 block and QT Long QT syndrome (LQTS) is linked to mutations in the prolongation (520 msec) in precordial lead V5 during ion channels, which can lead to disturbances in ventricu- sinus rhythm at 90 beats per minute (Fig. 1) and rhythm lar repolarization [1]. This condition puts patients at risk strip while walking (Fig. -

Splicing Repression Is a Major Function of Tdp-43 in Motor Neurons
SPLICING REPRESSION IS A MAJOR FUNCTION OF TDP-43 IN MOTOR NEURONS: THERAPEUTIC IMPLICATIONS FOR AMYOTROPHIC LATERAL SCLEROSIS by Aneesh N. Donde A thesis submitted to Johns Hopkins University in conformity with the requirements for the degree of Doctor of Philosophy Baltimore, Maryland May, 2018 Title: Splicing Repression is a Major Function of Tdp-43 in Motor Neurons Ph.D Dissertator: Aneesh N. Donde Ph.D Advisor: Philip C. Wong, Ph.D Abstract Nuclear depletion of TDP-43, an RNA binding protein which serves to protect the transcriptome by repressing aberrant splicing, may underlie neurodegeneration in amyotrophic lateral sclerosis (ALS). As multiple functions have been ascribed to TDP-43, whether splicing repression is its major role in motor neurons – that may be compromised in ALS – remains to be established. Here, we show that TDP-43 mediated splicing repression is central to the physiology of motor neurons. To validate TDP-43 mediated splicing repression as a therapeutic target, an AAV9-mediated gene delivery approach was employed to deliver a chimeric protein comprised of the N-terminal RNA recognition domain of TDP-43 fused to an unrelated splicing repressor (RAVER1) to mice lacking TDP-43 in motor neurons. This strategy allowed long-term expression of the repressor without any untoward effects, delayed the onset and slowed the progression of disease, and extended survival. In treated mice, evidence of aberrant splicing was markedly decreased and accompanied by amelioration of motor neuron loss. These findings establish that splicing repression is a principal role of TDP-43 in motor neurons and support the idea that loss of TDP-43-mediated splicing repression represents a key pathogenic mechanism underling motor neuron loss, validating a novel mechanism-based therapeutic strategy for ALS. -

South Australian Government Boards and Committees Information As at 30 June 2020
OFFICIAL South Australian Government Boards and Committees Information As at 30 June 2020 OFFICIAL OFFICIAL South Australian Government Boards and Committees As at 30 June 2020 Introduction This is the 24th annual report to Parliament of consolidated South Australian Government board and committee information1. The report sets out the membership and remuneration arrangements of 196 part-time government boards and committees as at 30 June 2020 in order of ministerial portfolio. The information has been sourced from the Boards and Committees Information System (BCIS), a database administered by the Department of the Premier and Cabinet, following extensive consultation with all ministerial offices and stakeholder agencies. Definition of boards and committees in the report The boards and committees included in this report are those which are: • established by or under an Act of Parliament of South Australia (generally excluding the Local Government Act 1999) and have a majority of members appointed by either a minister or the Governor; or • established by a minister or legal instrument such as a constitution or charter, have a majority of members appointed by a minister, and have at least one member in receipt of remuneration. The report should not be considered to be a complete listing of all government boards and committees. 1 Note: The 2019 report, which was the 23rd such report, was incorrectly published as the 22nd report. This error dates to the 2014 annual report, which was incorrectly published as the 17th report, resulting in all subsequent reports being incorrectly labelled. This error has been corrected in the copies of the report available on the DPC website Page 2 of 9 OFFICIAL OFFICIAL Highlights Number of boards and committees 196 boards and committees are identified in the 2020 report. -

In Re Network Associates, Inc. Securities Litigation 00-CV-4849
MC66N Rejected or Ineligible Claimants Page 1 of 251 MC66N138 NETWORK ASSOCIATES, INC. II SECUR REPS 13-Jun-05 11:50 AM Reason Deemed Claim Number Name City State Ineligible 10559 WATKINS, JOYCE E ATLANTA GA FATAL LINE 12498 1199 HEALTH CARE EMP CHICAGO IL NO LOSS 7784 16105 PIMCO IARORCIH NEW YORK NY NO LOSS 2259 20 UIT FUNDS NEWYORK NY INTRINSICALLY INELIGIBLE 2109830 33 WEST CLINTON AVEN TOMS RIVER NJ NO LOSS 9826 3M ERIP TRUST PITTSBURGH PA FATAL LINE 8466 4 Z'S INVESTMENTS PA WATERFORD MI NO LOSS 1080 45 INDIANA HOSPITOL PHILA PA NO LOSS 8779 777-H F II 1990 STLM DETROIT MI NO LOSS 2093721 800 PRE-EMPTION ROAD GENEVA NY NO LOSS 2247 91325 CANADA INC NO LOSS 8375 A DIAMOND FAMILY TRU LOS ANGELES CA NO LOSS 2065510 A F & N CIRAMELLA RE DAYTON OH NO LOSS 2029983 A R W SUPPORT TRUST TAYLORVILLE IL INTRINSICALLY INELIGIBLE 12103 AAA MI RESTRUCTURING CHICAGO IL NO LOSS 2093159 AAOF ENDOWMENT FUND SAINT LOUIS MO NO LOSS 2095827 AARON, BRIAN & CAROL BETHESDA MD FATAL LINE 2091134 ABAD, MARIO & NAVATA WEST ORANGE NJ NO LOSS 2027060 ABBAMONTE, MICHAEL & SOLON OH INTRINSICALLY INELIGIBLE 2101711 ABBETT, DEANNA L SAN JOSE CA FATAL LINE 2055264 ABDELQADER, MAHMUD S COLLINSVILLE IL NO LOSS 2030513 ABEGGLEN, CHERYL ROS CHADRON NE INTRINSICALLY INELIGIBLE 4336 ABEL, DAVID MELVILLE NY NO LOSS 2015464 ABEL, KEVIN KEW GARDENS NY NO LOSS 2020708 ABEL, STEPHEN ONEIDA NY NO LOSS 8542 ABEUTT, BETWORT MERRICK NY NO LOSS 2127049 ABLEY, PETER & VIBEK NO LOSS 6756 ABN AMRO N QUINCY MA INTRINSICALLY INELIGIBLE 4204 ABN AMRO SMALL CAP F CHICAGO IL NO LOSS 1214 -
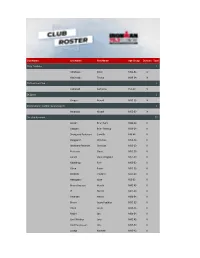
Club Name Last Name First Name Age Group Division Total 3City
Club Name Last Name First Name Age Group Division Total 3City Triathlon 2 Adolfsson Peter M50-54 IV Hekimoglu Tayfun M30-34 IV 3D Triathlon Club 1 Campbell Caitriona F55-59 V 3K Sport 1 Gregec Nenad M35-39 IV 338 Småland Triathlon & Multisport 1 Bergqvist Mikael M55-59 IV /tri club denmark 37 Jensen Brian Nors M40-44 II Ottosen Brian Peldrup M30-34 II Skovgaard Pedersen Camilla F40-44 II Daugaard Christian M50-54 II Wolthers-Petersen Christian M35-39 II Petersen Claus M55-59 II Larsen Claus Wiegand M55-59 II Moeldrup Finn M55-59 II Olsen Frank M55-59 II Ellekilde Frederik M25-29 II Hebsgaard Gitte F55-59 II Bruun Axelsen Henrik M45-49 II If Henrik M65-69 II Petersen Henrik M50-54 II Bruun Jacob Fuglkjær M35-39 II Olsen Jacob M30-34 II Reichl Jan M50-54 II Lind-Winther Jens M45-49 II Hvid Rasmussen Jim M55-59 II Luxhøi Kenneth M45-49 II Fischer Lars M45-49 II Vogelius Lasse M30-34 II Kjær Olesen Malene F30-34 II Vergroesen Marcus M30-34 II Andersen Martin M40-44 II Orby Martin Falck M30-34 II Mortensen Mathias Loft M25-29 II Andersen Mathias Meier M25-29 II Rolsted Michael M45-49 II Haakonsen Nick M40-44 II Jakobsen Nikolaj M25-29 II skjoldemose nikolaj M30-34 II Rosenlund Nino M45-49 II Larsen Peter M55-59 II Pauly Rune M45-49 II Kousgaard Sune M25-29 II Gammelvind Thomas M40-44 II Aalborg Triathlonklub 1 Kvist-Ibsen Anna F18-24 V Aegir 3 13 Jónsdóttir Dagný F30-34 V Jónsdóttir Erna Hlif F40-44 V Thorarinsson Finnbogi M55-59 V Jónsdóttir Guðrún F45-49 V Karlsdottir Inga Dagmar F45-49 V Poulsen Ingi M40-44 V Jónsson Jón Orri M25-29 V Steinadottir Kristin L. -

The Role of Parkin and Mitophagy in Acetaminophen and Alcohol-Induced Liver Injuries
The Role of Parkin and Mitophagy in Acetaminophen and Alcohol-induced Liver Injuries By Jessica A. Williams Submitted to the graduate degree program in Pharmacology, Toxicology, and Therapeutics and the Graduate Faculty of the University of Kansas in partial fulfillment of the requirements for the degree of Doctor of Philosophy. ________________________________ Chairperson Dr. Wen-Xing Ding, PhD ________________________________ Dr. Hartmut Jaeschke, PhD ________________________________ Dr. Michele Pritchard, PhD ________________________________ Dr. Udayan Apte, PhD ________________________________ Dr. Benyi Li, MD, PhD Date Defended: May 28th, 2015 The Dissertation Committee for Jessica A. Williams certifies that this is the approved version of the following dissertation: The Role of Parkin and Mitophagy in Acetaminophen and Alcohol-induced Liver Injuries ________________________________ Chairperson Dr. Wen-Xing Ding Date approved: June 8th, 2015 ii Abstract Acetaminophen (APAP) is the leading cause of acute liver failure in the United States, and alcoholic liver disease (ALD) is a worldwide health problem that claims two million lives per year. Currently, the only cure for either disease is liver transplantation in severe disease states. Therefore, new therapeutic options for treatment of these liver diseases are greatly needed. To develop new therapeutic options, the mechanisms involved in APAP and alcohol-induced liver toxicities must be better understood. We previously demonstrated that autophagy was protective against both APAP and alcohol-induced liver injuries by removing damaged mitochondria by mitophagy, which is a selective form of autophagy specific for mitochondria. However, the mechanisms for induction of mitophagy in the liver are unknown. Parkin is an E3 ubiquitin ligase that is well known to be required for mitophagy induction in mammalian cell models after mitochondrial depolarization. -

THE ANNALS of APPLIED STATISTICS
ISSN 1932-6157 (print) ISSN 1941-7330 (online) THE ANNALS of APPLIED STATISTICS AN OFFICIAL JOURNAL OF THE INSTITUTE OF MATHEMATICAL STATISTICS Articles Refining cellular pathway models using an ensemble of heterogeneous data sources ALEXANDER M. FRANKS,FLORIAN MARKOWETZ AND EDOARDO M. AIROLDI 1361 Statistical shape analysis of simplified neuronal trees ADAM DUNCAN,ERIC KLASSEN AND ANUJ SR I VA S TAVA 1385 TPRM: Tensor partition regression models with applications in imaging biomarker detection . MICHELLE F. MIRANDA,HONGTU ZHU AND JOSEPH G. IBRAHIM 1422 Complex-valued time series modeling for improved activation detection in fMRI studies...........DANIEL W. ADRIAN,RANJAN MAITRA AND DANIEL B. ROWE 1451 Optimal multilevel matching using network flows: An application to a summer reading intervention.........................SAMUEL D. PIMENTEL,LINDSAY C. PAGE, MATTHEW LENARD AND LUKE KEELE 1479 Topological data analysis of single-trial electroencephalographic signals YUAN WANG,HERNANDO OMBAO AND MOO K. CHUNG 1506 Joint significance tests for mediation effects of socioeconomic adversity on adiposity via epigenetics...............................................YEN-TSUNG HUANG 1535 Adaptive-weight burden test for associations between quantitative traits and genotype data withcomplexcorrelations...........XIAOWEI WU,TING GUAN,DAJIANG J. LIU, LUIS G. LEÓN NOVELO AND DIPANKAR BANDYOPADHYAY 1558 Bayesian aggregation of average data: An application in drug development SEBASTIAN WEBER,ANDREW GELMAN,DANIEL LEE,MICHAEL BETANCOURT, AKI VEHTARI AND AMY RACINE-POON 1583 BayCount: A Bayesian decomposition method for inferring tumor heterogeneity using RNA-Seq counts . FANGZHENG XIE,MINGYUAN ZHOU AND YANXUN XU 1605 Exploring the conformational space for protein folding with sequential Monte Carlo.......................SAMUEL W. K. WONG,JUN S. LIU AND S. C. -

Banking Act Unclaimed Money As at 31 December 2007
Commonwealth of Australia Gazette No. ASIC 40A/08, Wednesday, 21 May 2008 Published by ASIC ASIC Gazette Contents Banking Act Unclaimed Money as at 31 December 2007 RIGHTS OF REVIEW Persons affected by certain decisions made by ASIC under the Corporations Act 2001 and the other legislation administered by ASIC may have rights of review. ASIC has published Regulatory Guide 57 Notification of rights of review (RG57) and Information Sheet ASIC decisions – your rights (INFO 9) to assist you to determine whether you have a right of review. You can obtain a copy of these documents from the ASIC Digest, the ASIC website at www.asic.gov.au or from the Administrative Law Co-ordinator in the ASIC office with which you have been dealing. ISSN 1445-6060 (Online version) Available from www.asic.gov.au ISSN 1445-6079 (CD-ROM version) Email [email protected] © Commonwealth of Australia, 2008 This work is copyright. Apart from any use permitted under the Copyright Act 1968, all rights are reserved. Requests for authorisation to reproduce, publish or communicate this work should be made to: Gazette Publisher, Australian Securities and Investment Commission, GPO Box 9827, Melbourne Vic 3001 ASIC GAZETTE Commonwealth of Australia Gazette ASIC 40A/08, Wednesday, 21 May 2008 Banking Act Unclaimed Money Page 2 of 463 Specific disclaimer for Special Gazette relating to Banking Unclaimed Monies The information in this Gazette is provided by Authorised Deposit-taking Institutions to ASIC pursuant to the Banking Act (Commonwealth) 1959. The information is published by ASIC as supplied by the relevant Authorised Deposit-taking Institution and ASIC does not add to the information. -

Creighton Law Review
CREIGHTON LAW REVIEW Vol. 50, No. 2 2016-2017 SCHOOL OF LAW CREIGHTON UNIVERSITY OMAHA, NEBRASKA CREIGHTON LAW REVIEW BOARD OF EDITORS CLAIRE E. WILKA Editor in Chief PETER M. LANGDON Executive Editor TYLER S. SEALS BENJAMIN DEAVER KATIE M. MATEJKA Senior Lead Articles Editor Lead Articles Editor Lead Articles Editor MICHAEL SALLOUM SEAN T. NAKAMOTO SHANNON M. BEHM Lead Articles Editor Research Editor Student Articles Editor CAMERON OAKLEY FINKE NOAH GLOVER MORGAN L. KREISER Student Articles Editor Student Articles Editor Student Articles Editor EDITORIAL STAFF MARK HANNA LUKE HENKENIUS JEREMY FONTAIN ASSOCIATE STAFF RYAN P. CALLEY TAYLOR ANTHONY CLAPP MARIA A. COHEE ScoTT M. ECKEL LAUREL FREEMYER RACHEL M. LEE RILEY J. MCCORMICK CHRISTIAN H. MIRCH JULIE M. RYAN LExY K. SCHUMAN TIMOTHY A. SNYDER COURTNEY E. SOLMA AMANDA C. SWISHER RHYS J. WILLIAMS GENERAL STAFF PAUL JAMES BLAZEK BRENNAN R. BLOCK BLAKE MILLER YOUNSUNG PARK MIKAYLA L. TRAVERS DANIEL WILLIS FACULTY ADVISOR BusiNEss MANAGER NICHOLAS A. MIRKAY III DIANE KRILEY CREIGHTON LAW REVIEW TRIBUTES 50 YEARS OF THE CREIGHTON LAW REVIEW VOLUME 42 ............................ Darin L. Whitmer 179 VOLUME 49 ........................... Spencer R. Murphy '181 ARTICLES "MIRROR, MIRROR, ON THE WALL . .": REFLECTIONS ON FAIRNESS AND HOUSING IN THE OMAHA-COUNCIL BLUFFS REGION .... Palma Joy Strand 183 A CASE FOR THE DUE PROCESS RIGHT TO A SPEEDY EXTRADITION .............. Artemio Rivera 249 BATMAN AND TWo VERY LARGE JARS OF MAYONNAISE: THE LOOMING CLASH OF DAILY FANTASY SPORTS AND TRIBAL GAMING ...... Brett Wessels 295 JUSTICE AND BOUNDED MORAL RATIONALITY IN BANKRUPTCY .................................... Jooho Lee 333 NOTES DOES ACTUAL INNOCENCE ACTUALLY MATTER? WHY THE SCHLUP ACTUAL INNOCENCE GATEWAY REQUIRES NEWLY PRESENTED, RELIABLE EVIDENCE ......................... -

TEACHER Vol 13.Pdf
University “St. Kliment Ohridski“ Faculty of Education - Bitola TEACHER International Journal of Education Bitola, 2017 Publisher Faculty of Education - Bitola Dean prof. Valentina Gulevska, PhD. Executive and Editor-in-chief Prof. Ljupco Kevereski, PhD, Macedonia Editorail Board Academisian Grozdanka Gojkov, Serbia Academisian Marjan Blazic, Slovenia Prof. Milan Matijevik, PhD, Croatia Prof. Svetlana Kurtesh, PhD, England Prof. Danimir Mandic, PhD, Serbia Prof. Danijela Kostadinovic, PhD, Serbia Prof. Jasmina Starc, PhD, Slovenia Prof. Mojca Juriševič, PhD, Slovenia Prof. Anton Ilica, PhD, Romania Prof. Eva Soradova, PhD, Slovakia Prof. Lazar Stošić, PhD, Serbia Prof. Alla Belousova, PhD, Russia Prof. Irina Abakumova, PhD, Russia Prof. Tom Jovanovski, PhD, USA Prof. Jove D. Talevski, PhD, Macedonia Prof. Zlatko Zoglev, PhD, Macedonia Prof. Dobri Petrovski, PhD, Macedonia Prof. Metodija Stojanovski, PhD, Macedonia Cover, Technical & Computer support Josif Petrovski, Macedonia CIP - Cataloging in Publication, National and University Library "St. Kliment Ohridski" - Skopje. TEACHER: Journal of the Faculty of Education - Bitola / [Editorial Board Acad. Grozdanka Gojkov ... ] Year XV, No. 1 (2017) -. - Bitola: Faculty of Education, 2017 -. - 29 cm., 160 p. Unspecified ISSN 1857- 8888 (online) University “St. Kliment Ohridski“ - Bitola, Macedonia Faculty of Education - Bitola, Macedonia Address: Faculty of Education ul “Vasko karangelevski“ b.b. 7000 Bitola, Macedonia Tel/Fax. ++ 389 47 253 652; 203 385 With the opinion of the Ministry of Culture no. 07-2699/2 from 15.04.1998, for the journal "Teacher" is paid preferential tax rate. In accordance with Article 20, paragraph 8 of the VAT Law (Official Gazette 44/99), for the journal "Teacher” is paid a tax of 5%. The journal has no commercial nature.