2/19/08 CE 573 Lab 2-Microbial Enumeration Introduction There Are
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Product List 2008/09
Product List 2008/09 Part of Thermo Fisher Scientific Oxoid Chromogenic Media … Sheer Brilliance™ to reflect this, the names of products are being changed so that the current range now includes: Brilliance Bacillus cereus Agar Brilliance Candida Agar Brilliance E. coli/coliform Agar Brilliance E. coli/coliform Selective Agar Brilliance Enterobacter sakazakii Agar (DFI) Brilliance Listeria Agar Brilliance UTI Agar Brilliance UTI Clarity Agar Brilliance Salmonella Agar The chromogens used in our Brilliance range of chromogenic media ensure that colony colours are vivid, allowing rapid, easy differentiation and presumptive identification of the organism in question. So remember Brilliance – chromogenic media that delivers clearly visible answers on a single culture plate. Contents 1 Culture Media & Supplements 2 Supplementary Reagents 20 Antibiotic Single Supplements 21 Biochemical Reagents 22 Laboratory Preparations & Biological Extracts 22 Veggietones 24 Bagged Media 25 Dip Slides 25 Atmosphere Generation 26 AGS 26 Traditional Systems 27 Food Testing 28 DuPont Qualicon 29 BAX® System, Test and Components 29 Culture Media for the BAX System 30 RiboPrinter® Microbial Characterisation System 31 Lateral Flow System Range 31 Blood Culture 32 Signal 32 Wampole Isolator® 32 Antimicrobial Susceptibility Testing 33 M.I.C.Evaluator Strips 33 aura image 34 AST Accessories 34 Antimicrobial Susceptibility Testing Discs 35 Miscellaneous 38 Sundries 38 Publications 38 Diagnostics 39 Biochemical I.D. 39 Toxin Detection Kits 40 Agglutination Tests 41 Enzyme Immunoassays 42 Immunofluorescence Assays 43 Lateral Flow Assays 44 Environmental Monitoring and Process Simulation 45 Oxoid Air Sampler 45 Environmental Monitoring Swabs 45 Prepared Media 45 Dehydrated Media 45 Process Simulation 46 2 Culture Media and Supplements A wide range of general purpose, enrichment, selective and differential Anaerobe Basal Broth media, diluents and supplements a medium for general growth of anaerobes NB: information provided here is intended only as an aid to purchasing. -

View Our Full Water Sampling Vials Product Offering
TABLE OF CONTENTS Environmental Monitoring 1 Sample Prep and Dilution 8 Dehydrated Culture Media - Criterion™ 12 Prepared Culture Media 14 Chromogenic Media - HardyCHROM™ 18 CompactDry™ 20 Quality Control 24 Membrane Filtration 25 Rapid Tests 26 Lab Supplies/Sample Collection 27 Made in the USA Headquarters Distribution Centers 1430 West McCoy Lane Santa Maria, California Santa Maria, CA 93455 Olympia, Washington 800.266.2222 : phone Salt Lake City, Utah [email protected] Phoenix, Arizona HardyDiagnostics.com Dallas, Texas Springboro, Ohio Hardy Diagnostics has a Quality Lake City, Florida Management System that is certified Albany, New York to ISO 13485 and is a FDA licensed Copyright © 2020 Hardy Diagnostics Raleigh, North Carolina medical device manufacturer. Environmental Monitoring Hardy Diagnostics offers a wide selection of quality products intended to help keep you up to date with regulatory compliance, ensure the efficacy of your cleaning protocol, and properly monitor your environment. 800.266.2222 [email protected] HardyDiagnostics.com 1 Environmental Monitoring Air Sampling TRIO.BAS™ Impact Air Samplers introduced the newest generation of microbial air sampling. These ergonomically designed instruments combine precise air sampling with modern connectivity to help you properly assess the air quality of your laboratory and simplify your process. MONO DUO Each kit includes: Each kit includes: TRIO.BAS™ MONO air sampler, induction TRIO.BAS™ DUO air sampler, battery battery charger and cable, aspirating charger -

WHO Global Foodborne Infections Network
WHO Global Foodborne Infections Network (formerly WHO Global Salm-Surv) "A WHO network building capacity to detect, control and prevent foodborne and other enteric infections from farm to table” Laboratory Protocol “Isolation of Salmonella spp. From Food and Animal Faeces ” 5th Ed. June 2010 1 IMPORTANT NOTES: 1) This procedure is based on the ISO protocol: 6579:2002 “Microbiology of food and animal feeding stuffs -- Horizontal method for the detection of Salmonella spp.”4. This protocol is intended to provide guidance for the testing of suspect food items/ animal faecal specimens identified via foodborne disease surveillance programmes. Regulatory agencies (Ministries of Health, Agriculture, Commerce, etc) have specific testing requirements, different from this protocol, which much be used to test samples collected for regulatory testing (example: import/export or product recall). Prior to performing any official, legal, or regulatory testing, the reader should confirm the appropriate protocol through consult with in-country regulatory authorities. 2) This protocol is intended only to be used on food samples and animal faeces. This protocol should not be used for the testing of human faeces. Foreword: Infections due to Salmonella spp. remain a global problem. These infections may cause significant morbidity and mortality both in humans and production animals as well as considerable economic losses. Salmonella spp. are typically transmitted among humans and animals via a fecal-oral route, usually through the consumption of contaminated food or water. Timely identification and serotyping of Salmonella from clinical specimens facilitates outbreak detection and patient management while prompt and accurate detection of Salmonella spp. in contaminated food or water provides an opportunity to prevent the contaminated food from entering the food supply. -
Nutrient Broth (N7519)
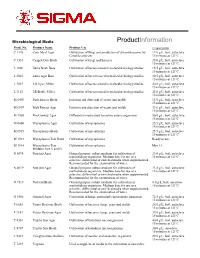
Microbiological Media ProductInformation Prod. No. Product Name Product Use Preparation C 1176 Corn Meal Agar Cultivation of fungi and production of chlamydospores by 17.0 g/L, boil, autoclave Candida albicans 15 minutes at 121ºC. C 1551 Czapek Dox Broth Cultivation of fungi and bacteria 35.0 g/L, boil, autoclave 15 minutes at 121ºC. L 1900 Luria Broth Base Cultivation of bacteria used in molecular biology studies. 15.5 g/L, boil, autoclave 15 minutes at 121ºC. L 2025 Luria Agar Base Cultivation of bacteria used in molecular biology studies. 30.5 g/L, boil, autoclave 15 minutes at 121ºC. L 3027 LB Agar, Miller Cultivation of bacteria used in molecular biology studies. 40.0 g/L; boil, autoclave 15 minutes at 121ºC. L 3152 LB Broth, Miller Cultivation of bacteria used in molecular biology studies. 25.0 g/L; boil, autoclave 15 minutes at 121ºC. M 6409 Malt Extract Broth Isolation and detection of yeasts and molds 15.0 g/L, boil, autoclave 15 minutes at 121ºC. M 6907 Malt Extract Agar Isolation and detection of yeasts and molds 33.6 g/L, boil, autoclave 15 minutes at 121ºC. M 7408 MacConkey Agar Differential media used to isolate enteric organisms 50.0 g/L; boil, autoclave 15 minutes at 121ºC M 0660 Mycoplasma Agar Cultivation of mycoplasma 35.5 g/L; boil, autoclave 15 minutes at 121ºC M 0535 Mycoplasma Broth Cultivation of mycoplasma 25.5 g/L; boil, autoclave 15 minutes at 121ºC M 1539 Mycoplasma Test Broth Cultivation of mycoplasma Ready to use. M 1914 Mycoplasma Test Cultivation of mycoplasma Mix 1:1. -

Bacterial Cultures Care Guide SCIENTIFIC Introduction Use the Following Guide to Learn How to Properly Care for Bacterial Cultures
Bacterial Cultures Care Guide SCIENTIFIC Introduction Use the following guide to learn how to properly care for bacterial cultures. BIO FAX! Safety Precautions After use, agar plates will likely contain viable microbes. Although the bacteria are not likely to be pathogenic, do not open the plates unnecessarily. Use sterile techniques at all times when handling bacterial cultures. When plates are done being used, they should be autoclaved or opened under a 10% bleach solution and soaked for at least one hour. Bleach solution is a corrosive liquid that may dis- color clothing and cause skin burns. Avoid contact of bleach with heat, acids and organic materials; chlorine gas will be generated. Wear chemical splash goggles, chemical-resistant gloves, and a chemical-resistant apron. Wash hands thoroughly with soap and water before leaving the laboratory. Please follow all laboratory safety guidelines. Culturing and Maintenance Prior to receiving bacterial cultures it is helpful to be aware of the required conditions for the particular strain of bacteria purchased. Each species has specific conditions that are necessary for optimal growth. Refer to the Bacterial Cultures Table below for incubation temperature and the appropriate general medium. Bacterial Cultures Table Description (Genus species) Incubation Temperature† Medium Bacillus cereus (20–35) 30 °C Nutrient agar Bacillus mycoides 30 °C Nutrient agar Bacillus megaterium (25–35) 30 °C Nutrient agar Bacillus subtilis (25–35) 25–30 °C Nutrient agar Enterobacter aerogenes (30–37) 30 °C Nutrient -

LAB 81.7 Eng.Pdf (1.188Mb)
WORLD HEALTH ORGANIZATION LAB/81. 7 ORGANISATION MONDIALE DE LA SANTE 8RIGHh\L: ENGLISH CA) L.'"( GUIDELINES FOR THE LABORATORY DIAGNOSIS OF DI~IA,.. tf.A.~A '( by Mr R. Brooks Head Medical Laboratory Scientific Officer Public Health Laboratory Swansea 1 Glamorgan, United Kingdom CONTENTS 1. INTRODUCTION . 2 2. HEALTH SIGNIFICANCE OF DIPHTHERIA 2 3. THE IMPORTANT ROLE OF THE LABORATORY IN THE DIAGNOSIS OF DIPHTHERIA 2 4. PROCEDURES FOR THE ISOLATION AND IDENTIFICATION OF CORYNEBACTERIUM DIPHTHERIAE 3 4.1 Collection and transport of specimens (swabs) 4 4.1.1 Materials required for swabbing and transport of swabs 5 4.1. 2 Throat (swab) specimens . 5 4.1. 3 Nasopharyngeal (swab) specimens . 5 4.1. 4 Skin diphtheria and other diphtheritic lesions 5 4.2 Processing of collected (swab) specimens for the isolation of C. diphtheriae. 5 4.2.1 Minimum culture media necessary for the isolation of C. diphtheriae. 6 4.2.2 Minimum requirements for the inoculation and incubation of culture media . 7 4.2.3 Primary plating of (swab) specimens for the isolation of C. diphtheriae . • . 7 4.3 Examination of cultures for the presence of C. diphtheriae 7 4.3.1 Examination of BA plate 7 4.3.2 Examination of TBA plate 9 4.4 Identification of C. diphtheriae 9 4.4.1 Method of obtaining pure cultures 9 4.4.2 Toxigenicity testing of C. diphtheriae 10 4.4.3 Biochemical testing of C. diphtheriae 14 4.4.4 Biotyping of C. diphtheriae 17 APPENDIX - REAGENTS AND CULTURE MEDIA PREPARATION ...... 0 0 0 ••••• 0 0 ••••••• 0 •••••••• 0 0 0 •• 0 The issue of this document does not constitute Ce document ne constitue pas une publication. -

The Microbial Community of Kitchen Sponges: Experimental Study Investigating Bacterial Number, Resistance and Transfer
Digital Commons @ Assumption University Honors Theses Honors Program 2019 The Microbial Community of Kitchen Sponges: Experimental Study Investigating Bacterial Number, Resistance and Transfer Sydney Knoll Assumption College Follow this and additional works at: https://digitalcommons.assumption.edu/honorstheses Part of the Life Sciences Commons Recommended Citation Knoll, Sydney, "The Microbial Community of Kitchen Sponges: Experimental Study Investigating Bacterial Number, Resistance and Transfer" (2019). Honors Theses. 54. https://digitalcommons.assumption.edu/honorstheses/54 This Honors Thesis is brought to you for free and open access by the Honors Program at Digital Commons @ Assumption University. It has been accepted for inclusion in Honors Theses by an authorized administrator of Digital Commons @ Assumption University. For more information, please contact [email protected]. The Microbial Community of Kitchen Sponges: Experimental Study Investigating Bacterial Number, Resistance and Transfer Sydney Knoll Faculty Supervisor: Aisling Dugan, Ph.D. Department of Biological and Physical Sciences A Thesis Submitted to Fulfill the Requirements of the Honors Program at Assumption College Spring 2019 Acknowledgments I would like to express my deepest appreciation to Professor Aisling Dugan for serving as my honors advisor, mentor and role model in my life. I am incredibly grateful for her continued support and the countless number of times she has read and edited my thesis. This work would not have been possible without her. I would also like to acknowledge the Assumption College Honors Program and the Department of Biological and Physical Sciences for their support and financial assistance they provided me throughout this research. Finally, I would like to thank my parents for being there for me every step of the way through college, including this thesis. -

BBL Nutrient Agar
BBL™ Nutrient Agar ! L007481 • Rev. 08 • July 2006 " QUALITY CONTROL PROCEDURES I INTRODUCTION Nutrient Agar is a general-purpose medium for the cultivation of a wide variety of bacterial organisms. II PERFORMANCE TEST PROCEDURE 1. Liquefy Nutrient Agar in A tubes by heating in boiling water. Cool to 45–50°C and pour into Petri dishes and allow to firm up for at least 30 min. 2. Streak the plates using 0.01 mL calibrated loops in order to obtain isolated colonies. For tubed slants, inoculate the agar surfaces with a loopful of inoculum. Use 10-1 dilutions of 18- to 24-h Trypticase™ Soy Broth cultures of the organisms listed below. 3. Incubate plates or tubes in an aerobic atmosphere at 35 ± 2°C. Caps should be loosened on the tubed media. 4. Examine plates or tubes after 18–24 h for amount of growth. 5. Expected Results Organisms ATCC™ Recovery Pseudomonas aeruginosa 10145 Moderate to heavy growth, green pigmentation *Shigella flexneri 12022 Moderate to heavy growth *Staphylococcus aureus 25923 Moderate to heavy growth, cream to gold colonies *Recommended organism strain for User Quality Control. III ADDITIONAL QUALITY CONTROL 1. Examine tubes as described under “Product Deterioration.” 2. Visually examine representative tubes to assure that any existing physical defects will not interfere with use. 3. Incubate uninoculated representative tubes at 20–25°C and 30–35°C and examine after 7 days for microbial contamination. PRODUCT INFORMATION IV INTENDED USE Nutrient Agar is used for the cultivation of bacteria and for the enumeration of organisms in water, sewage, feces and other materials. -

Prepared Culture Media
PREPARED CULTURE MEDIA 030220SG PREPARED CULTURE MEDIA Made in the USA AnaeroGRO™ DuoPak A 02 Bovine Blood Agar, 5%, with Esculin 13 AnaeroGRO™ DuoPak B 02 Bovine Blood Agar, 5%, with Esculin/ AnaeroGRO™ BBE Agar 03 MacConkey Biplate 13 AnaeroGRO™ BBE/PEA 03 Bovine Selective Strep Agar 13 AnaeroGRO™ Brucella Agar 03 Brucella Agar with 5% Sheep Blood, Hemin, AnaeroGRO™ Campylobacter and Vitamin K 13 Selective Agar 03 Brucella Broth with 15% Glycerol 13 AnaeroGRO™ CCFA 03 Brucella with H and K/LKV Biplate 14 AnaeroGRO™ Egg Yolk Agar, Modifi ed 03 Buffered Peptone Water 14 AnaeroGRO™ LKV Agar 03 Buffered Peptone Water with 1% AnaeroGRO™ PEA 03 Tween® 20 14 AnaeroGRO™ MultiPak A 04 Buffered NaCl Peptone EP, USP 14 AnaeroGRO™ MultiPak B 04 Butterfi eld’s Phosphate Buffer 14 AnaeroGRO™ Chopped Meat Broth 05 Campy Cefex Agar, Modifi ed 14 AnaeroGRO™ Chopped Meat Campy CVA Agar 14 Carbohydrate Broth 05 Campy FDA Agar 14 AnaeroGRO™ Chopped Meat Campy, Blood Free, Karmali Agar 14 Glucose Broth 05 Cetrimide Select Agar, USP 14 AnaeroGRO™ Thioglycollate with Hemin and CET/MAC/VJ Triplate 14 Vitamin K (H and K), without Indicator 05 CGB Agar for Cryptococcus 14 Anaerobic PEA 08 Chocolate Agar 15 Baird-Parker Agar 08 Chocolate/Martin Lewis with Barney Miller Medium 08 Lincomycin Biplate 15 BBE Agar 08 CompactDry™ SL 16 BBE Agar/PEA Agar 08 CompactDry™ LS 16 BBE/LKV Biplate 09 CompactDry™ TC 17 BCSA 09 CompactDry™ EC 17 BCYE Agar 09 CompactDry™ YMR 17 BCYE Selective Agar with CAV 09 CompactDry™ ETB 17 BCYE Selective Agar with CCVC 09 CompactDry™ YM 17 -

NRM Nutritional Requirements and Media Learning Objectives the Student Will Use Aseptic Techniques in the Safe Inoculation of Various Forms of Media
NRM Nutritional Requirements and Media Learning Objectives The student will Use aseptic techniques in the safe inoculation of various forms of media. Follow oral and written instructions and manage time in the lab efficiently. Apply correct terminology regarding microbiological techniques, instruments, microbial growth, media types and forms when making observations. Correctly perform various inoculation techniques. Background/Theory Organisms can be identified according to the source of carbon they use for metabolism as well as their energy source. The prefixes auto- (“self”) and hetero- (“other”) refer to the origins of the carbon sources various organisms can use. Organisms that convert inorganic carbon dioxide (CO2) into organic carbon compounds are autotrophs. Plants and cyanobacteria are well-known examples of autotrophs. Conversely, heterotrophs rely on more complex organic carbon compounds as nutrients; these are provided to them initially by autotrophs. Many organisms, ranging from humans to many prokaryotes, including the well-studied Escherichia coli, are heterotrophic. (OpenStax CNX, 2018) This exercise investigates the nutritional requirements of some heterotrophic bacteria. Heterotrophs need an organic source of carbon for ATP generation and for anabolic (growth) activities. Carbohydrates, proteins and fats are all organic compounds. In addition, nitrogen is needed for building amino acids which form proteins. Nitrogen can come from organic sources (proteins, for example) or inorganic sources. Some heterotrophs can manufacture many complex molecules from inorganic and small organic molecules. Others rely on many large, more complex molecules being available in the environment. The more ready-made, complex organic compounds an organism requires of the environment, the more fastidious it is. In this course you will encounter many types of growth media. -

2020/2021 Microbiology Products Catalog
2020/2021 Microbiology Products Catalog Clinical • Food & Beverage • Pharmaceutical • Environmental • Veterinary HOW TO ORDER Contact details Technical Support helpline Thermo Fisher Specialty Diagnostics Ltd Our web site is intended to make it easier for you to ask Wade Road questions or raise issues with us. Simply go to www. Basingstoke thermofisher.com/microbiology, and choose the most Hampshire appropriate option in “Contact Us”. RG24 8PW UK Technical enquiries can also be made by telephone, by fax or online: Tel: +44 (0) 1256 694288 Tel: +44 (0) 1256 694238 Email: [email protected] Email: [email protected] Web: www.thermofisher.com/microbiology UK Office Hours: Mon-Fri 08.00 - 17.00 Sat & Sun closed Contents Antimicrobial Susceptibility Testing 6 Diagnostics 57 Antimicrobial Susceptibility Testing Discs 6 Biochemical Identification 57 Antifungal Susceptibility Testing Discs 8 Immunological Tests 59 AST Media 8 Blood and Serum Tests 61 AST Accessories 8 Toxin Detection Kits 62 Sensititre ID/AST System 9 Agglutination Tests 62 Sensititre Plates 9 Enzyme Immunoassays 65 Sensititre Equipment 10 ELISA Accessories 66 Sensititre Accessories 10 Food Allergen ELISA Assays 66 Sensititre Media 10 Immunofluorescence Assays 67 Lateral Flow Assays 68 Incubators 11 Reagents and Stains 68 Atmosphere Generation Systems 12 Laboratory Supplies 69 AGS 12 Molecular Products 71 Blood Culture 13 Sample Prep for Molecular and Non-Molecular Workflows 71 Food Molecular Detection and Quantification -

Anaerobe Agar
Thermo Scientific Microbiology Prepared media selection guide For the isolation, identification, differentiation and susceptibility testing of microorganisms Table of Contents Anaerobe Agar 4 Antimicrobial Susceptibility Testing (AST) Agars 6 Bi-plates 9 Blood Agars 14 Brilliance Chromogenic Media (Clinical) 16 Brilliance Chromogenic Media (Food) 21 Diluents, Water & Peptones 23 Dip-Slides 24 General Purpose Media 25 Pharmaceutical Media 29 ReadyBags 32 Water Testing 33 Culture Media byby Organism Type Aeromonas 36 ListeriaListeria 5252 Bacillus cereus 366 MycoplasmaMycoplasma / Ureaplasma 52 Bordetella 37 PasteurellaPasteurella 5353 Burkholderia cepaciaacia 37 Pseudomonas 53 Campylobacter 388 SalmonellaSalmonella 5454 Clostridium specieses 38 Staphylococci / Streptococcii 5757 Coliforms / Escherichiarichia colii 4040 StaphylococcusStaphylococcus aureusaureus 5858 Corynebacteria 411 StreptococcusStreptococcus agalactiaeagalactiae 5959 Dermatophytes 41 Trichomonas 60 Escherichia coli O157 42 Vibrio 60 Enterobacteriaceae 43 Yeasts & Moulds 61 Enterococci 47 Yersinia 63 Gardnerella 48 Haemophilus & Neisseria 48 Helicobacter pylori 50 Lactobacilli / Bifidobacteria 50 Legionella 51 Bringing more to microbiology Highest levels of quality & consistency Our culture medium expertise and rigorous quality standards have made us a preferred supplier and trusted source of prepared media to laboratories around the world. With a full range of formulations and formats, our media products combine ease-of-use with accurate, reproducible performance. You can rest assured knowing the Thermo Scientific™ culture media that reaches your benchtop will provide optimal recovery and differentiation of organisms, for greater confidence in your results. Unmatched service & support When you choose Thermo Scientific products for your microbiology needs, consider it the start of a lifelong partnership. Whether you need assistance with protocols, product transitions or product troubleshooting, our team of experts is ready to help you.