Identification of Salmonella Species
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Produktkatalog 2015/2016
Produktkatalog 2015/2016 BD Life Sciences Diagnostic Systems Deutschland . Österreich . Schweiz BD stellt sich vor BD Lifeweltweit Sciences – BD – LifeBD MedicalSciences – BD Medical BD – weltweit BD Life Sciences – Biosciences BD Life Sciences – Diagnostic Systems BD, eines der führenden Medizintech- Biosciences bedient den gesamten Diagnostic Systems ist Ihr Experte und nologie-Unternehmen, hat es sich zur Bereich der biologischen und medizi- erfahrener Partner für die klinische Aufgabe gemacht, in Zusammenar- nischen Forschung und bietet die und industrielle mikrobiologische Diag- beit mit seinen Kunden und Partnern größte Auswahl an innovativen Gerä- nostik. den gegenwärtigen und zukünftigen ten und Reagenzien für die zelluläre Umfassende Konzepte und daran ge- Herausforderungen bei der Gesund- Analyse an. knüpfte Diagnostika, Softwarelösun- heitsversorgung von Menschen auf Ein breites Spektrum an Analyzern gen und Dienstleistungen führen zu der ganzen Welt zu begegnen. und Sortern für die Durchflusszyto- gesteigerter Effizienz im Labor und Mit innovativen Lösungen optimiert metrie, verbunden mit kompetenter zu schnelleren verwertbaren Ergeb- BD die Arzneimittelverabreichung, Beratung und Service, ermöglichen nissen. verbessert die Diagnose von Infek- die passgenaue Lösung für Ihr Labor. Unser klinischer Fokus liegt in der Prä- tionskrankheiten und Krebs, unter- Unser Produktangebot und unsere vention und Diagnose von Infektions- stützt die Behandlung von Diabetes Kompetenz machen uns zum interes- krankheiten sowie des Zervixkarzinoms. und bringt die Zellforschung voran. santen Partner für Forschungs- und Für den klinischen Bereich bietet Diag- Rund 30.000 BD-Mitarbeiterinnen Routinelabors, für die Industrie sowie nostic Sytems integrierte Lösungen und Mitarbeiter in 50 Ländern setzen für die wachsende Zahl von bio- und auf den Gebieten Tuberkulose, Sep- sich dafür ein, unser Unternehmens- gentechnologischen Einrichtungen. -

Salmonella Species
MICROBIAL FACTSHEET SERIES ISSUE NO. 1 | SEPTEMBER 2011 Salmonella species 1. What are Salmonella spp.? Salmonella spp. are a group of bacteria which reside in the intestinal tract of human beings and warm blooded animals and are capable of causing disease. They are the second most common cause of bacterial foodborne illness in Ireland (Campylobacter spp. is the most frequent cause). They are facultative anaerobic Gram- negative rods. Salmonella spp. are members of the Enterobacteriaceae group. The genus Salmonella contains 2 species: • Salmonella enterica • Salmonella bongori Salmonella enterica is an important agent of foodborne illness. This species is sub-classified into 6 subspecies of which S. enterica subspecies enterica is the most important for human health. The genus Salmonella can be subdivided into more than 2,400 serotypes. Salmonella enterica subsp. enterica serotype Typhimurium (S. Typhimurium) and Salmonella enterica subsp. enterica serotype Enteritidis (S. Enteritidis) are the most frequently isolated serotypes in humans in Ireland. Serotypes are further sub- divided by their resistance to bacteriophages (phage types or lystotypes), antibiotics or heavy metals; their biochemical characteristics (biovars or biotypes) or their sensitivity to or production of bacteriocins. 2. Growth and Survival Characteristics Table 1. Factors affecting the growth of Salmonella spp. COndITIOnS MInIMuM OpTIMuM MAxIMuM Temperature (ºC) 5.2* 35 – 43 46.2 pH 3.8 7 – 7.5 9.5 Water activity (aw) 0.94 0.99 >0.99 * Most serotypes fail to grow at < 7ºC Salmonella spp. are not particularly heat resistant and most serotypes are killed by normal cooking conditions, i.e. cooking to a core temperature of 75ºC instantaneously or an equivalent time temperature combination, e.g. -

OXOID MANUAL PRELIMS 16/6/06 12:18 Pm Page 1
OXOID MANUAL PRELIMS 16/6/06 12:18 pm Page 1 The OXOID MANUAL 9th Edition 2006 Compiled by E. Y. Bridson (substantially revised) (former Technical Director of Oxoid) Price: £50 OXOID MANUAL PRELIMS 16/6/06 12:18 pm Page 2 The OXOID MANUAL 9th Edition 2006 Compiled by E. Y. Bridson (substantially revised) (former Technical Director of Oxoid) 9th Edition 2006 Published by OXOID Limited, Wade Road, Basingstoke, Hampshire RG24 8PW, England Telephone National: 01256 841144 International: +44 1256 841144 Email: [email protected] Facsimile National: 01256 463388 International: +44 1256 463388 Website http://www.oxoid.com OXOID SUBSIDIARIES AROUND THE WORLD AUSTRALIA DENMARK NEW ZEALAND Oxoid Australia Pty Ltd Oxoid A/S Oxoid NZ Ltd 20 Dalgleish Street Lunikvej 28 3 Atlas Place Thebarton, Adelaide DK-2670 Greve, Denmark Mairangi Bay South Australia 5031, Australia Tel: 45 44 97 97 35 Auckland 1333, New Zealand Tel: 618 8238 9000 or Fax: 45 44 97 97 45 Tel: 00 64 9 478 0522 Tel: 1 800 331163 Toll Free Email: [email protected] NORWAY Fax: 618 8238 9060 or FRANCE Oxoid AS Fax: 1 800 007054 Toll Free Oxoid s.a. Nils Hansen vei 2, 3 etg Email: [email protected] 6 Route de Paisy BP13 0667 Oslo BELGIUM 69571 Dardilly Cedex, France PB 6490 Etterstad, 0606 Oxoid N.V./S.A. Tel: 33 4 72 52 33 70 Oslo, Norway Industriepark, 4E Fax: 33 4 78 66 03 76 Tel: 47 23 03 9690 B-9031 Drongen, Belgium Email: [email protected] Fax: 47 23 09 96 99 Tel: 32 9 2811220 Email: [email protected] GERMANY Fax: 32 9 2811223 Oxoid GmbH SPAIN Email: [email protected] Postfach 10 07 53 Oxoid S.A. -

Salmonella Bongori Provides Insights Into the Evolution of the Salmonellae
Salmonella bongori Provides Insights into the Evolution of the Salmonellae Maria Fookes1", Gunnar N. Schroeder2", Gemma C. Langridge1", Carlos J. Blondel3, Caterina Mammina4, Thomas R. Connor1, Helena Seth-Smith1, Georgios S. Vernikos1, Keith S. Robinson2, Mandy Sanders1, Nicola K. Petty1, Robert A. Kingsley1, Andreas J. Ba¨umler5, Sean-Paul Nuccio5,Ine´s Contreras3, Carlos A. Santiviago3, Duncan Maskell6, Paul Barrow7, Tom Humphrey8, Antonino Nastasi9, Mark Roberts10, Gad Frankel2, Julian Parkhill1, Gordon Dougan1, Nicholas R. Thomson1* 1 Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, United Kingdom, 2 Centre for Molecular Microbiology and Infection, Division of Cell and Molecular Biology, Imperial College London, London, United Kingdom, 3 Departamento de Bioquı´mica y Biologı´a Molecular, Facultad de Ciencias Quı´micas y Farmace´uticas, Universidad de Chile, Santiago, Chile, 4 Dept. Sciences for Health Promotion ‘‘G. D’Alessandro’’, University of Palermo, Palermo, Italy, 5 Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, Davis, California, United State of America, 6 Department of Veterinary Medicine, University of Cambridge, Cambridge, United Kingdom, 7 School of Veterinary Medicine and Science, University of Nottingham, Sutton Bonington, Leicestershire, United Kingdom, 8 National Centre for Zoonosis Research, University of Liverpool, Leahurst Campus, Neston, Wirral, United Kingdom, 9 Dipartimento di Sanita` Publica, Universita` di Firenze, Italy, 10 Institute of Comparative Medicine, Faculty of Veterinary Medicine, University of Glasgow, Glasgow, United Kingdom Abstract The genus Salmonella contains two species, S. bongori and S. enterica. Compared to the well-studied S. enterica there is a marked lack of information regarding the genetic makeup and diversity of S. -

Laboratory Diagnosis of Shigella and Salmonella Infections *
Bull. Org. mond. Sante) 1959, 21, Bull. Wid Hlth Org. 247-277 Laboratory Diagnosis of Shigella and Salmonella Infections * E. HORMAECHE, M.D.' & C. A. PELUFFO, M.D.2 In recent years a great deal of work has been We are aware that experienced bacteriologists devoted to the study of enteric diseases and, in will prefer some of the methods they are using to consequence, many methods have been developed the ones we recommend. This study is not in- for the isolation of their most common agents, tended for them, however, but rather as a guide to Shigella, Salmonella and some pathogenic Esche- people who are just starting work on enteric diseases richia, while at the same time new tests have been diagnosis. For more detailed information, we devised to differentiate the " groups " of the Entero- recommend the books of Kauffmann (1954) and bacteriaceae family, so that the beginner, especially, Edwards & Ewing (1955), of which we have made will be faced with the difficulty of selecting from ample use. among them those to be used. Investigation of enteric pathogens will sometimes Each author has his preferences according to his be the ultimate purpose, as for the public health own experience, and it is regrettable that up to now officer, who is primarily interested in finding them no serious effort has been made to standardize for the potential danger they represent to the com- techniques, which would save a lot of time and dis- munity. In clinical medicine, however, the role couraging failures for the bacteriologist unex- played by the bacteriologist does not end there. -

Medios En Placas Preparadas
Medios de cultivo Hemoderivados, Medios en placas preparadas Sheep Blood in Alsevers Solution Iso-Sensitest Agar (Placa profunda) 25 mL SR0053B 10 x 90 mm PO0779A Iso-Sensitest Agar with Chocolated Horse Blood Sheep Blood Defibrinated 10 x 90 mm PB0147A 25 mL SR0051B Iso-Sensitest Agar with Horse Blood 10 x 90 mm PB0146A Medios en placas preparadas Iso-Sensitest Agar with Horse Blood and 20mg/L NAD 10 x 90 mm PB0378A Agares sangre Iso-Sensitest Agar con sangre de caballo lisada Blood Agar Base No. 2 with Horse Blood 10 x 90 mm PB0145A 10 x 90 mm PB0114A Iso-Sensitest Agar with Sheep Blood Blood Agar Base No. 2 with Sheep Blood 10 x 90 mm PB5003A 10 x 90 mm PB0115A Mueller Hinton Agar Columbia Agar with Horse Blood 10 x 90 mm PO5007A 10 x 90 mm PB0122A Mueller-Hinton Agar with 2% Salt Columbia Agar with Sheep Blood 10 x 90 mm PO0799A^ 10 x 90 mm PB5008A 10 x 90 mm PO5139A+ Columbia Agar with Sheep Blood PLUS Mueller-Hinton Agar 10 x 90 mm PB0123A*^ 10 x 90 mm PO0152A^ 10 x 90 mm PB5039A+ 10x120mm PO1191S Columbia Agar with Chocolated Horse Blood Mueller-Hinton Agar with 5% Sheep Blood 10 x 90 mm PB0124A 10 x 90 mm PB0431A^ 10 x 90 mm PB5007A+ Agares generales para anaerobios Mueller Hinton Agar with Horse Blood Thermo ScientificTM A.R.I.A.TM with Horse Blood 10 x 90 mm PB1229A^ 10 x 90 mm PB1243A 10 x 90 mm PB5303A+ A.R.I.A. -

Product List 2008/09
Product List 2008/09 Part of Thermo Fisher Scientific Oxoid Chromogenic Media … Sheer Brilliance™ to reflect this, the names of products are being changed so that the current range now includes: Brilliance Bacillus cereus Agar Brilliance Candida Agar Brilliance E. coli/coliform Agar Brilliance E. coli/coliform Selective Agar Brilliance Enterobacter sakazakii Agar (DFI) Brilliance Listeria Agar Brilliance UTI Agar Brilliance UTI Clarity Agar Brilliance Salmonella Agar The chromogens used in our Brilliance range of chromogenic media ensure that colony colours are vivid, allowing rapid, easy differentiation and presumptive identification of the organism in question. So remember Brilliance – chromogenic media that delivers clearly visible answers on a single culture plate. Contents 1 Culture Media & Supplements 2 Supplementary Reagents 20 Antibiotic Single Supplements 21 Biochemical Reagents 22 Laboratory Preparations & Biological Extracts 22 Veggietones 24 Bagged Media 25 Dip Slides 25 Atmosphere Generation 26 AGS 26 Traditional Systems 27 Food Testing 28 DuPont Qualicon 29 BAX® System, Test and Components 29 Culture Media for the BAX System 30 RiboPrinter® Microbial Characterisation System 31 Lateral Flow System Range 31 Blood Culture 32 Signal 32 Wampole Isolator® 32 Antimicrobial Susceptibility Testing 33 M.I.C.Evaluator Strips 33 aura image 34 AST Accessories 34 Antimicrobial Susceptibility Testing Discs 35 Miscellaneous 38 Sundries 38 Publications 38 Diagnostics 39 Biochemical I.D. 39 Toxin Detection Kits 40 Agglutination Tests 41 Enzyme Immunoassays 42 Immunofluorescence Assays 43 Lateral Flow Assays 44 Environmental Monitoring and Process Simulation 45 Oxoid Air Sampler 45 Environmental Monitoring Swabs 45 Prepared Media 45 Dehydrated Media 45 Process Simulation 46 2 Culture Media and Supplements A wide range of general purpose, enrichment, selective and differential Anaerobe Basal Broth media, diluents and supplements a medium for general growth of anaerobes NB: information provided here is intended only as an aid to purchasing. -

View Our Full Water Sampling Vials Product Offering
TABLE OF CONTENTS Environmental Monitoring 1 Sample Prep and Dilution 8 Dehydrated Culture Media - Criterion™ 12 Prepared Culture Media 14 Chromogenic Media - HardyCHROM™ 18 CompactDry™ 20 Quality Control 24 Membrane Filtration 25 Rapid Tests 26 Lab Supplies/Sample Collection 27 Made in the USA Headquarters Distribution Centers 1430 West McCoy Lane Santa Maria, California Santa Maria, CA 93455 Olympia, Washington 800.266.2222 : phone Salt Lake City, Utah [email protected] Phoenix, Arizona HardyDiagnostics.com Dallas, Texas Springboro, Ohio Hardy Diagnostics has a Quality Lake City, Florida Management System that is certified Albany, New York to ISO 13485 and is a FDA licensed Copyright © 2020 Hardy Diagnostics Raleigh, North Carolina medical device manufacturer. Environmental Monitoring Hardy Diagnostics offers a wide selection of quality products intended to help keep you up to date with regulatory compliance, ensure the efficacy of your cleaning protocol, and properly monitor your environment. 800.266.2222 [email protected] HardyDiagnostics.com 1 Environmental Monitoring Air Sampling TRIO.BAS™ Impact Air Samplers introduced the newest generation of microbial air sampling. These ergonomically designed instruments combine precise air sampling with modern connectivity to help you properly assess the air quality of your laboratory and simplify your process. MONO DUO Each kit includes: Each kit includes: TRIO.BAS™ MONO air sampler, induction TRIO.BAS™ DUO air sampler, battery battery charger and cable, aspirating charger -

Molecular Typing and Evolutionary Relationships of Salmonella Enterica Serovar Typhi
Molecular typing and evolutionary relationships of Salmonella enterica serovar Typhi Sophie Octavia School of Biotechnology and Biomolecular Sciences The University of New South Wales Australia A thesis submitted for the degree of Doctor of Philosophy March 2008 PLEASE TYPE 1.3.1 THE UNIVERSITY OF NEW SOUTH WALES Thesis/Dissertation Sheet Surname or Family name: Octavia First name: Sophie Other name/s: PhD in Microbiology Abbreviation for degree as given in the University calendar: School: Biotechnology and Biomolecular Sciences Faculty: Science Title: Molecular typing and evolutionary relationships of Salmonella enterica serovar Typhi Abstract 350 words maximum: (PLEASE TYPE) The evolutionary relationship between Salmonella enterica serovar Typhi, other typhoid-like enteric fever causing serovars and 10 non- Typhoid serovars from S. enterica subspecies I, could not be determined by comparative nucleotide sequences of six genes. Phylogenetic analyses of the dataset showed that the genes of interest underwent frequent recombination, suggesting a low level of clonality within subspecies I of S. enterica. To establish the evolutionary relationships within serovar Typhi, genome-wide Single Nucleotide Polymorphism (SNP) was explored as a marker for both typing purposes and phylogenetic analysis. Thirty eight SNPs were typed in 73 global Typhi isolates, including 18 isolates expressing the special flagellar antigen z66, using restriction enzyme digestion method. The isolates were differentiated into 23 SNP profiles and grouped into four distinct clusters. The z66 isolates were divided into four SNP profiles and were all grouped into one cluster, suggesting a single origin. An alternative SNP typing method using the hairpin real time PCR assay was investigated to type four additional SNPs, termed as biallelic polymorphisms (BiP). -

Shigella Dysenteriae Type 1
Guidelines for the control of shigellosis, including epidemics due to Shigella dysenteriae type 1 World Health Organization TABLE OF CONTENTS i WHO Library Cataloguing-in-Publication Data World Health Organization. Guidelines for the control of shigellosis, including epidemics due to Shigella dysenteriae 1. 1.Dysentery, Bacillary - prevention and control 2.Shigella dysenteriae - pathogenicity 3.Disease outbreaks - prevention and control 4.Guidelines I.Title. ISBN 92 4 159233 0 NLM Classification: WC 282) © World Health Organization 2005 All rights reserved. Publications of the World Health Organization can be obtained from WHO Press, World Health Organization, 20 Avenue Appia, 1211 Geneva 27, Switzerland (tel: +41 22 791 2476; fax: +41 22 791 4857; email: [email protected]). Requests for permission to reproduce or translate WHO publications – whether for sale or for noncommercial distribution – should be addressed to WHO Press, at the above address (fax: +41 22 791 4806; email: [email protected]). The designations employed and the presentation of the material in this publication do not imply the expression of any opinion whatsoever on the part of the World Health Organization concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. Dotted lines on maps represent approximate border lines for which there may not yet be full agreement. The mention of specific companies or of certain manufacturers’ products does not imply that they are endorsed or recommended by the World Health Organization in preference to others of a similar nature that are not mentioned. Errors and omissions excepted, the names of proprietary products are distinguished by initial capital letters. -

WHO Global Foodborne Infections Network
WHO Global Foodborne Infections Network (formerly WHO Global Salm-Surv) "A WHO network building capacity to detect, control and prevent foodborne and other enteric infections from farm to table” Laboratory Protocol “Isolation of Salmonella spp. From Food and Animal Faeces ” 5th Ed. June 2010 1 IMPORTANT NOTES: 1) This procedure is based on the ISO protocol: 6579:2002 “Microbiology of food and animal feeding stuffs -- Horizontal method for the detection of Salmonella spp.”4. This protocol is intended to provide guidance for the testing of suspect food items/ animal faecal specimens identified via foodborne disease surveillance programmes. Regulatory agencies (Ministries of Health, Agriculture, Commerce, etc) have specific testing requirements, different from this protocol, which much be used to test samples collected for regulatory testing (example: import/export or product recall). Prior to performing any official, legal, or regulatory testing, the reader should confirm the appropriate protocol through consult with in-country regulatory authorities. 2) This protocol is intended only to be used on food samples and animal faeces. This protocol should not be used for the testing of human faeces. Foreword: Infections due to Salmonella spp. remain a global problem. These infections may cause significant morbidity and mortality both in humans and production animals as well as considerable economic losses. Salmonella spp. are typically transmitted among humans and animals via a fecal-oral route, usually through the consumption of contaminated food or water. Timely identification and serotyping of Salmonella from clinical specimens facilitates outbreak detection and patient management while prompt and accurate detection of Salmonella spp. in contaminated food or water provides an opportunity to prevent the contaminated food from entering the food supply. -
Nutrient Broth (N7519)
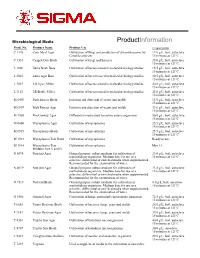
Microbiological Media ProductInformation Prod. No. Product Name Product Use Preparation C 1176 Corn Meal Agar Cultivation of fungi and production of chlamydospores by 17.0 g/L, boil, autoclave Candida albicans 15 minutes at 121ºC. C 1551 Czapek Dox Broth Cultivation of fungi and bacteria 35.0 g/L, boil, autoclave 15 minutes at 121ºC. L 1900 Luria Broth Base Cultivation of bacteria used in molecular biology studies. 15.5 g/L, boil, autoclave 15 minutes at 121ºC. L 2025 Luria Agar Base Cultivation of bacteria used in molecular biology studies. 30.5 g/L, boil, autoclave 15 minutes at 121ºC. L 3027 LB Agar, Miller Cultivation of bacteria used in molecular biology studies. 40.0 g/L; boil, autoclave 15 minutes at 121ºC. L 3152 LB Broth, Miller Cultivation of bacteria used in molecular biology studies. 25.0 g/L; boil, autoclave 15 minutes at 121ºC. M 6409 Malt Extract Broth Isolation and detection of yeasts and molds 15.0 g/L, boil, autoclave 15 minutes at 121ºC. M 6907 Malt Extract Agar Isolation and detection of yeasts and molds 33.6 g/L, boil, autoclave 15 minutes at 121ºC. M 7408 MacConkey Agar Differential media used to isolate enteric organisms 50.0 g/L; boil, autoclave 15 minutes at 121ºC M 0660 Mycoplasma Agar Cultivation of mycoplasma 35.5 g/L; boil, autoclave 15 minutes at 121ºC M 0535 Mycoplasma Broth Cultivation of mycoplasma 25.5 g/L; boil, autoclave 15 minutes at 121ºC M 1539 Mycoplasma Test Broth Cultivation of mycoplasma Ready to use. M 1914 Mycoplasma Test Cultivation of mycoplasma Mix 1:1.