WHO Global Foodborne Infections Network
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Superficieibacter Electus Gen. Nov., Sp. Nov., an Extended-Spectrum β
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Digital Commons@Becker Washington University School of Medicine Digital Commons@Becker Open Access Publications 2018 Superficieibacter electus gen. nov., sp. nov., an extended-spectrum β-lactamase possessing member of the enterobacteriaceae family, isolated from Intensive Care Unit surfaces Robert F. Potter Washington University School of Medicine in St. Louis Alaric W. D-Souza Washington University School of Medicine in St. Louis Meghan A. Wallace Washington University School of Medicine in St. Louis Angela Shupe Washington University School of Medicine in St. Louis Sanket Patel Washington University School of Medicine in St. Louis See next page for additional authors Follow this and additional works at: https://digitalcommons.wustl.edu/open_access_pubs Recommended Citation Potter, Robert F.; D-Souza, Alaric W.; Wallace, Meghan A.; Shupe, Angela; Patel, Sanket; Gul, Danish; Kwon, Jennie H.; Beatty, Wandy; Andleeb, Saadia; Burnham, Carey-Ann D.; and Dantas, Gautam, ,"Superficieibacter electus gen. nov., sp. nov., an extended- spectrum β-lactamase possessing member of the enterobacteriaceae family, isolated from Intensive Care Unit surfaces." Frontiers in Microbiology.9,. 1629. (2018). https://digitalcommons.wustl.edu/open_access_pubs/7020 This Open Access Publication is brought to you for free and open access by Digital Commons@Becker. It has been accepted for inclusion in Open Access Publications by an authorized administrator of Digital Commons@Becker. For more information, please contact [email protected]. Authors Robert F. Potter, Alaric W. D-Souza, Meghan A. Wallace, Angela Shupe, Sanket Patel, Danish Gul, Jennie H. Kwon, Wandy Beatty, Saadia Andleeb, Carey-Ann D. -

Product List 2008/09
Product List 2008/09 Part of Thermo Fisher Scientific Oxoid Chromogenic Media … Sheer Brilliance™ to reflect this, the names of products are being changed so that the current range now includes: Brilliance Bacillus cereus Agar Brilliance Candida Agar Brilliance E. coli/coliform Agar Brilliance E. coli/coliform Selective Agar Brilliance Enterobacter sakazakii Agar (DFI) Brilliance Listeria Agar Brilliance UTI Agar Brilliance UTI Clarity Agar Brilliance Salmonella Agar The chromogens used in our Brilliance range of chromogenic media ensure that colony colours are vivid, allowing rapid, easy differentiation and presumptive identification of the organism in question. So remember Brilliance – chromogenic media that delivers clearly visible answers on a single culture plate. Contents 1 Culture Media & Supplements 2 Supplementary Reagents 20 Antibiotic Single Supplements 21 Biochemical Reagents 22 Laboratory Preparations & Biological Extracts 22 Veggietones 24 Bagged Media 25 Dip Slides 25 Atmosphere Generation 26 AGS 26 Traditional Systems 27 Food Testing 28 DuPont Qualicon 29 BAX® System, Test and Components 29 Culture Media for the BAX System 30 RiboPrinter® Microbial Characterisation System 31 Lateral Flow System Range 31 Blood Culture 32 Signal 32 Wampole Isolator® 32 Antimicrobial Susceptibility Testing 33 M.I.C.Evaluator Strips 33 aura image 34 AST Accessories 34 Antimicrobial Susceptibility Testing Discs 35 Miscellaneous 38 Sundries 38 Publications 38 Diagnostics 39 Biochemical I.D. 39 Toxin Detection Kits 40 Agglutination Tests 41 Enzyme Immunoassays 42 Immunofluorescence Assays 43 Lateral Flow Assays 44 Environmental Monitoring and Process Simulation 45 Oxoid Air Sampler 45 Environmental Monitoring Swabs 45 Prepared Media 45 Dehydrated Media 45 Process Simulation 46 2 Culture Media and Supplements A wide range of general purpose, enrichment, selective and differential Anaerobe Basal Broth media, diluents and supplements a medium for general growth of anaerobes NB: information provided here is intended only as an aid to purchasing. -

Food Microbiology
Food Microbiology Food Water Dairy Beverage Online Ordering Available Food, Water, Dairy, & Beverage Microbiology Table of Contents 1 Environmental Monitoring Contact Plates 3 Petri Plates 3 Culture Media for Air Sampling 4 Environmental Sampling Boot Swabs 6 Environmental Testing Swabs 8 Surface Sanitizers 8 Hand Sanitation 9 Sample Preparation - Dilution Vials 10 Compact Dry™ 12 HardyCHROM™ Chromogenic Culture Media 15 Prepared Media 24 Agar Plates for Membrane Filtration 26 CRITERION™ Dehydrated Culture Media 28 Pathogen Detection Environmental With Monitoring Contact Plates Baird Parker Agar Friction Lid For the selective isolation and enumeration of coagulase-positive staphylococci (Staphylococcus aureus) on environmental surfaces. HardyCHROM™ ECC 15x60mm contact plate, A chromogenic medium for the detection, 10/pk ................................................................................ 89407-364 differentiation, and enumeration of Escherichia coli and other coliforms from environmental surfaces (E. coli D/E Neutralizing Agar turns blue, coliforms turn red). For the enumeration of environmental organisms. 15x60mm plate contact plate, The media is able to neutralize most antiseptics 10/pk ................................................................................ 89407-354 and disinfectants that may inhibit the growth of environmental organisms. Malt Extract 15x60mm contact plate, Malt Extract is recommended for the cultivation and 10/pk ................................................................................89407-482 -

Prepared Culture Media
PREPARED CULTURE MEDIA 121517SS PREPARED CULTURE MEDIA Made in the USA AnaeroGRO™ DuoPak A 02 Bovine Blood Agar, 5%, with Esculin 13 AnaeroGRO™ DuoPak B 02 Bovine Blood Agar, 5%, with Esculin/ AnaeroGRO™ BBE Agar 03 MacConkey Biplate 13 AnaeroGRO™ BBE/PEA 03 Bovine Selective Strep Agar 13 AnaeroGRO™ Brucella Agar 03 Brucella Agar with 5% Sheep Blood, Hemin, AnaeroGRO™ Campylobacter and Vitamin K 13 Selective Agar 03 Brucella Broth with 15% Glycerol 13 AnaeroGRO™ CCFA 03 Brucella with H and K/LKV Biplate 14 AnaeroGRO™ Egg Yolk Agar, Modified 03 Buffered Peptone Water 14 AnaeroGRO™ LKV Agar 03 Buffered Peptone Water with 1% AnaeroGRO™ PEA 03 Tween® 20 14 AnaeroGRO™ MultiPak A 04 Buffered NaCl Peptone EP, USP 14 AnaeroGRO™ MultiPak B 04 Butterfield’s Phosphate Buffer 14 AnaeroGRO™ Chopped Meat Broth 05 Campy Cefex Agar, Modified 14 AnaeroGRO™ Chopped Meat Campy CVA Agar 14 Carbohydrate Broth 05 Campy FDA Agar 14 AnaeroGRO™ Chopped Meat Campy, Blood Free, Karmali Agar 14 Glucose Broth 05 Cetrimide Select Agar, USP 14 AnaeroGRO™ Thioglycollate with Hemin and CET/MAC/VJ Triplate 14 Vitamin K (H and K), without Indicator 05 CGB Agar for Cryptococcus 14 Anaerobic PEA 08 Chocolate Agar 15 Baird-Parker Agar 08 Chocolate/Martin Lewis with Barney Miller Medium 08 Lincomycin Biplate 15 BBE Agar 08 CompactDry™ SL 16 BBE Agar/PEA Agar 08 CompactDry™ LS 16 BBE/LKV Biplate 09 CompactDry™ TC 17 BCSA 09 CompactDry™ EC 17 BCYE Agar 09 CompactDry™ YMR 17 BCYE Selective Agar with CAV 09 CompactDry™ ETB 17 BCYE Selective Agar with CCVC 09 CompactDry™ YM 17 BCYE -

View Our Full Water Sampling Vials Product Offering
TABLE OF CONTENTS Environmental Monitoring 1 Sample Prep and Dilution 8 Dehydrated Culture Media - Criterion™ 12 Prepared Culture Media 14 Chromogenic Media - HardyCHROM™ 18 CompactDry™ 20 Quality Control 24 Membrane Filtration 25 Rapid Tests 26 Lab Supplies/Sample Collection 27 Made in the USA Headquarters Distribution Centers 1430 West McCoy Lane Santa Maria, California Santa Maria, CA 93455 Olympia, Washington 800.266.2222 : phone Salt Lake City, Utah [email protected] Phoenix, Arizona HardyDiagnostics.com Dallas, Texas Springboro, Ohio Hardy Diagnostics has a Quality Lake City, Florida Management System that is certified Albany, New York to ISO 13485 and is a FDA licensed Copyright © 2020 Hardy Diagnostics Raleigh, North Carolina medical device manufacturer. Environmental Monitoring Hardy Diagnostics offers a wide selection of quality products intended to help keep you up to date with regulatory compliance, ensure the efficacy of your cleaning protocol, and properly monitor your environment. 800.266.2222 [email protected] HardyDiagnostics.com 1 Environmental Monitoring Air Sampling TRIO.BAS™ Impact Air Samplers introduced the newest generation of microbial air sampling. These ergonomically designed instruments combine precise air sampling with modern connectivity to help you properly assess the air quality of your laboratory and simplify your process. MONO DUO Each kit includes: Each kit includes: TRIO.BAS™ MONO air sampler, induction TRIO.BAS™ DUO air sampler, battery battery charger and cable, aspirating charger -

Annual Conference 2018 Abstract Book
Annual Conference 2018 POSTER ABSTRACT BOOK 10–13 April, ICC Birmingham, UK @MicrobioSoc #Microbio18 Virology Workshop: Clinical Virology Zone A Presentations: Wednesday and Thursday evening P001 Rare and Imported Pathogens Lab (RIPL) turn around time (TAT) for the telephoned communication of positive Zika virus (ZIKV) PCR and serology results. Zaneeta Dhesi, Emma Aarons Rare and Imported Pathogens Lab, Public Health England, Salisbury, United Kingdom Abstract Background: RIPL introduced developmental assays for ZIKV PCR and serology on 18/01/16 and 10/03/16 respectively. The published ZIKV test TATs were 5 days for PCR and 7 days for serology. Methods: All ZIKV RNA positive, seroconversion and “probable” cases diagnosed at RIPL up until 31/05/17 were identified. For each case, the date on which the relevant positive sample was received, and the date on which it was telephoned out to the requestor was ascertained. The number of working days between these two dates was calculated. Results: ZIKV PCR - 151 ZIKV PCR positive results were identified, of which 4 samples were excluded because no TAT could be calculated. The mean TAT for the remaining 147 samples was 1.7 working days. Ninety percent of these results were telephoned within 3 or fewer days of the sample having been received. There was 1 sample where the TAT was above the 90th centile. ZIKV Serology - 147 seroconversion or “Probable” ZIKV cases diagnosed serologically were identified. The mean TAT for these samples was 2.5 working days. Ninety percent of these results were telephoned within 4 or fewer days of the sample having been received. -

Lysine Iron Agar (Lia)
LYSINE IRON AGAR (LIA) INTENDED USE Remel Lysine Iron Agar (LIA) is a solid medium recommended for use in qualitative procedures for differentiation of enteric gram-negative bacilli based on deamination or decarboxylation of lysine and production of hydrogen sulfide (H2S). SUMMARY AND EXPLANATION In 1961, Edwards and Fife described LIA for detection of lactose-fermenting Arizona strains implicated in outbreaks of food-borne disease.1 The differentiation of Arizona from Salmonella was necessary because both produce lysine decarboxylase rapidly and form large amounts of H2S and not all species cause illness in humans. Prior to the introduction of LIA, Triple Sugar Iron (TSI) Agar and Kligler Iron Agar (KIA) were used for detection of H2S. However, some H2S-positive enterics were found to produce acid levels in TSI Agar and KIA high enough to 2 3 suppress H2S production. In 1966, Johnson et al. expanded the use of LIA to include identification of all Enterobacteriaceae. It was determined that lysine-positive enteric gram-negative bacilli will produce detectable levels of H2S in LIA, even if not in KIA, because the alkaline pH that results from decarboxylation of lysine enhances precipitation of H2S. In later years, Ewing recommended the use of LIA in conjunction with TSI for the detection of enteric pathogens in routine examination of stools.4 PRINCIPLE Peptone and yeast extract provide nitrogen, amino acids, and vitamins necessary for bacterial growth. Dextrose is a source of fermentable carbohydrate and brom cresol purple is a pH indicator. Ferric ammonium citrate is an indicator of H2S production. If H2S is produced from sodium thiosulfate, it reacts with ferric ammonium citrate to form a black precipitate (ferrous sulfate) in the butt of the tube. -
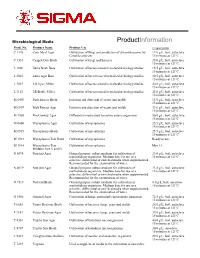
Nutrient Broth (N7519)
Microbiological Media ProductInformation Prod. No. Product Name Product Use Preparation C 1176 Corn Meal Agar Cultivation of fungi and production of chlamydospores by 17.0 g/L, boil, autoclave Candida albicans 15 minutes at 121ºC. C 1551 Czapek Dox Broth Cultivation of fungi and bacteria 35.0 g/L, boil, autoclave 15 minutes at 121ºC. L 1900 Luria Broth Base Cultivation of bacteria used in molecular biology studies. 15.5 g/L, boil, autoclave 15 minutes at 121ºC. L 2025 Luria Agar Base Cultivation of bacteria used in molecular biology studies. 30.5 g/L, boil, autoclave 15 minutes at 121ºC. L 3027 LB Agar, Miller Cultivation of bacteria used in molecular biology studies. 40.0 g/L; boil, autoclave 15 minutes at 121ºC. L 3152 LB Broth, Miller Cultivation of bacteria used in molecular biology studies. 25.0 g/L; boil, autoclave 15 minutes at 121ºC. M 6409 Malt Extract Broth Isolation and detection of yeasts and molds 15.0 g/L, boil, autoclave 15 minutes at 121ºC. M 6907 Malt Extract Agar Isolation and detection of yeasts and molds 33.6 g/L, boil, autoclave 15 minutes at 121ºC. M 7408 MacConkey Agar Differential media used to isolate enteric organisms 50.0 g/L; boil, autoclave 15 minutes at 121ºC M 0660 Mycoplasma Agar Cultivation of mycoplasma 35.5 g/L; boil, autoclave 15 minutes at 121ºC M 0535 Mycoplasma Broth Cultivation of mycoplasma 25.5 g/L; boil, autoclave 15 minutes at 121ºC M 1539 Mycoplasma Test Broth Cultivation of mycoplasma Ready to use. M 1914 Mycoplasma Test Cultivation of mycoplasma Mix 1:1. -

Bacterial Cultures Care Guide SCIENTIFIC Introduction Use the Following Guide to Learn How to Properly Care for Bacterial Cultures
Bacterial Cultures Care Guide SCIENTIFIC Introduction Use the following guide to learn how to properly care for bacterial cultures. BIO FAX! Safety Precautions After use, agar plates will likely contain viable microbes. Although the bacteria are not likely to be pathogenic, do not open the plates unnecessarily. Use sterile techniques at all times when handling bacterial cultures. When plates are done being used, they should be autoclaved or opened under a 10% bleach solution and soaked for at least one hour. Bleach solution is a corrosive liquid that may dis- color clothing and cause skin burns. Avoid contact of bleach with heat, acids and organic materials; chlorine gas will be generated. Wear chemical splash goggles, chemical-resistant gloves, and a chemical-resistant apron. Wash hands thoroughly with soap and water before leaving the laboratory. Please follow all laboratory safety guidelines. Culturing and Maintenance Prior to receiving bacterial cultures it is helpful to be aware of the required conditions for the particular strain of bacteria purchased. Each species has specific conditions that are necessary for optimal growth. Refer to the Bacterial Cultures Table below for incubation temperature and the appropriate general medium. Bacterial Cultures Table Description (Genus species) Incubation Temperature† Medium Bacillus cereus (20–35) 30 °C Nutrient agar Bacillus mycoides 30 °C Nutrient agar Bacillus megaterium (25–35) 30 °C Nutrient agar Bacillus subtilis (25–35) 25–30 °C Nutrient agar Enterobacter aerogenes (30–37) 30 °C Nutrient -

Superficieibacter Electus Gen. Nov., Sp. Nov., an Extended-Spectrum Β-Lactamase Possessing Member of the Enterobacteriaceae
ORIGINAL RESEARCH published: 20 July 2018 doi: 10.3389/fmicb.2018.01629 Superficieibacter electus gen. nov., sp. nov., an Extended-Spectrum β-Lactamase Possessing Member of the Enterobacteriaceae Family, Isolated From Intensive Care Unit Surfaces Robert F. Potter 1†, Alaric W. D’Souza 1†, Meghan A. Wallace 2, Angela Shupe 2, Sanket Patel 2, Danish Gul 3, Jennie H. Kwon 4, Wandy Beatty 5, Saadia Andleeb 3, Edited by: 2,5,6 1,2,5,7 Martin G. Klotz, Carey-Ann D. Burnham * and Gautam Dantas * Washington State University Tri-Cities, 1 The Edison Family Center for Genome Sciences and Systems Biology, Washington University in St. Louis School of United States Medicine, St. Louis, MO, United States, 2 Department of Pathology and Immunology, Washington University in St. Louis Reviewed by: School of Medicine, St. Louis, MO, United States, 3 Atta ur Rahman School of Applied Biosciences, National University of Sylvain Brisse, Sciences and Technology, Islamabad, Pakistan, 4 Division of Infectious Diseases, Washington University School of Medicine, Institut Pasteur, France St. Louis, MO, United States, 5 Department of Molecular Microbiology, Washington University in St. Louis School of Medicine, Awdhesh Kalia, St. Louis, MO, United States, 6 Department of Pediatrics, St. Louis Children’s Hospital, St. Louis, MO, United States, University of Texas MD Anderson 7 Department of Biomedical Engineering, Washington University in St. Louis, St. Louis, MO, United States Cancer Center, United States *Correspondence: Two Gram-negative bacilli strains, designated BP-1(T) and BP-2, were recovered from Carey-Ann D. Burnham [email protected] two different Intensive Care Unit surfaces during a longitudinal survey in Pakistan. -

Laboratory Methods for the Diagnosis of Epidemic Dysentery and Cholera Centers for Disease Control and Prevention Atlanta, Georgia 1999 WHO/CDS/CSR/EDC/99.8
WHO/CDS/CSR/EDC/99.8 Laboratory Methods for the Diagnosis of Epidemic Dysentery and Cholera Centers for Disease Control and Prevention Atlanta, Georgia 1999 WHO/CDS/CSR/EDC/99.8 Laboratory Methods for the Diagnosis of Epidemic Dysentery and Cholera Centers for Disease Control and Prevention Atlanta, Georgia 1999 This manual was prepared by the National Center for Infectious Diseases (NCID), Centers for Disease Control and Prevention (CDC), Atlanta, Georgia, USA, in cooperation with the World Health Organization Regional Office for Africa, (WHO/AFRO) Harare, Zimbabwe. Jeffrey P. Koplan, M.D., M.P.H., Director, CDC James M. Hughes, M.D., Director, NCID, CDC Mitchell L. Cohen, M.D., Director, Division of Bacterial and Mycotic Diseases, NCID, CDC Ebrahim Malek Samba, M.B.,B.S., Regional Director, WHO/AFRO Antoine Bonaventure Kabore, M.D., M.P.H., Director Division for Prevention and Control of Communicable Diseases, WHO/AFRO The following CDC staff members prepared this report: Cheryl A. Bopp, M.S. Allen A. Ries, M.D., M.P.H. Joy G. Wells, M.S. Production: J. Kevin Burlison, Graphics James D. Gathany, Photography Lynne McIntyre, M.A.L.S., Editor Cover: From top, Escherichia co//O157:H7 on sorbitol MacConkey agar, Vibrio cholerae O1 on TCBS agar, and Shige/la flexneri on xylose lysine desoxycholate agar. Acknowledgments Funding for the development of this manual was provided by the U.S. Agency for International Development, Bureau for Africa, Office of Sustainable Development. This manual was developed as a result of a joint effort by the World Health Organization Regional Office for Africa, WHO Headquarters, and the Centers for Disease Control and Prevention as part of the activities of the WHO Global Task Force on Cholera Control. -

LAB 81.7 Eng.Pdf (1.188Mb)
WORLD HEALTH ORGANIZATION LAB/81. 7 ORGANISATION MONDIALE DE LA SANTE 8RIGHh\L: ENGLISH CA) L.'"( GUIDELINES FOR THE LABORATORY DIAGNOSIS OF DI~IA,.. tf.A.~A '( by Mr R. Brooks Head Medical Laboratory Scientific Officer Public Health Laboratory Swansea 1 Glamorgan, United Kingdom CONTENTS 1. INTRODUCTION . 2 2. HEALTH SIGNIFICANCE OF DIPHTHERIA 2 3. THE IMPORTANT ROLE OF THE LABORATORY IN THE DIAGNOSIS OF DIPHTHERIA 2 4. PROCEDURES FOR THE ISOLATION AND IDENTIFICATION OF CORYNEBACTERIUM DIPHTHERIAE 3 4.1 Collection and transport of specimens (swabs) 4 4.1.1 Materials required for swabbing and transport of swabs 5 4.1. 2 Throat (swab) specimens . 5 4.1. 3 Nasopharyngeal (swab) specimens . 5 4.1. 4 Skin diphtheria and other diphtheritic lesions 5 4.2 Processing of collected (swab) specimens for the isolation of C. diphtheriae. 5 4.2.1 Minimum culture media necessary for the isolation of C. diphtheriae. 6 4.2.2 Minimum requirements for the inoculation and incubation of culture media . 7 4.2.3 Primary plating of (swab) specimens for the isolation of C. diphtheriae . • . 7 4.3 Examination of cultures for the presence of C. diphtheriae 7 4.3.1 Examination of BA plate 7 4.3.2 Examination of TBA plate 9 4.4 Identification of C. diphtheriae 9 4.4.1 Method of obtaining pure cultures 9 4.4.2 Toxigenicity testing of C. diphtheriae 10 4.4.3 Biochemical testing of C. diphtheriae 14 4.4.4 Biotyping of C. diphtheriae 17 APPENDIX - REAGENTS AND CULTURE MEDIA PREPARATION ...... 0 0 0 ••••• 0 0 ••••••• 0 •••••••• 0 0 0 •• 0 The issue of this document does not constitute Ce document ne constitue pas une publication.