Practical Microbiology for Secondary Schools
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Review Pili in Gram-Negative and Gram-Positive Bacteria – Structure
Cell. Mol. Life Sci. 66 (2009) 613 – 635 1420-682X/09/040613-23 Cellular and Molecular Life Sciences DOI 10.1007/s00018-008-8477-4 Birkhuser Verlag, Basel, 2008 Review Pili in Gram-negative and Gram-positive bacteria – structure, assembly and their role in disease T. Profta,c,* and E. N. Bakerb,c a School of Medical Sciences, Department of Molecular Medicine & Pathology, University of Auckland, Private Bag 92019, Auckland 1142 (New Zealand), Fax: +64-9-373-7492, e-mail: [email protected] b School of Biological Sciences, University of Auckland, Auckland (New Zealand) c Maurice Wilkins Centre for Molecular Biodiscovery, University of Auckland (New Zealand) Received 08 August 2008; received after revision 24 September 2008; accepted 01 October 2008 Online First 27 October 2008 Abstract. Many bacterial species possess long fila- special form of bacterial cell movement, known as mentous structures known as pili or fimbriae extend- twitching motility. In contrast, the more recently ing from their surfaces. Despite the diversity in pilus discovered pili in Gram-positive bacteria are formed structure and biogenesis, pili in Gram-negative bac- by covalent polymerization of pilin subunits in a teria are typically formed by non-covalent homopo- process that requires a dedicated sortase enzyme. lymerization of major pilus subunit proteins (pilins), Minor pilins are added to the fiber and play a major which generates the pilus shaft. Additional pilins may role in host cell colonization. be added to the fiber and often function as host cell This review gives an overview of the structure, adhesins. Some pili are also involved in biofilm assembly and function of the best-characterized pili formation, phage transduction, DNA uptake and a of both Gram-negative and Gram-positive bacteria. -

Exploring Small-Scale Chemostats to Scale up Microbial Processes: 3-Hydroxypropionic Acid Production in S
Lawrence Berkeley National Laboratory Recent Work Title Exploring small-scale chemostats to scale up microbial processes: 3-hydroxypropionic acid production in S. cerevisiae. Permalink https://escholarship.org/uc/item/43h8866j Journal Microbial cell factories, 18(1) ISSN 1475-2859 Authors Lis, Alicia V Schneider, Konstantin Weber, Jost et al. Publication Date 2019-03-11 DOI 10.1186/s12934-019-1101-5 Peer reviewed eScholarship.org Powered by the California Digital Library University of California Lis et al. Microb Cell Fact (2019) 18:50 https://doi.org/10.1186/s12934-019-1101-5 Microbial Cell Factories RESEARCH Open Access Exploring small-scale chemostats to scale up microbial processes: 3-hydroxypropionic acid production in S. cerevisiae Alicia V. Lis1, Konstantin Schneider1,2, Jost Weber1,3, Jay D. Keasling1,4,5,6,7, Michael Krogh Jensen1 and Tobias Klein1,2* Abstract Background: The physiological characterization of microorganisms provides valuable information for bioprocess development. Chemostat cultivations are a powerful tool for this purpose, as they allow defned changes to one single parameter at a time, which is most commonly the growth rate. The subsequent establishment of a steady state then permits constant variables enabling the acquisition of reproducible data sets for comparing microbial perfor- mance under diferent conditions. We performed physiological characterizations of a 3-hydroxypropionic acid (3-HP) producing Saccharomyces cerevisiae strain in a miniaturized and parallelized chemostat cultivation system. The physi- ological conditions under investigation were various growth rates controlled by diferent nutrient limitations (C, N, P). Based on the cultivation parameters obtained subsequent fed-batch cultivations were designed. Results: We report technical advancements of a small-scale chemostat cultivation system and its applicability for reliable strain screening under diferent physiological conditions, i.e. -

Detection of Acid-Producing Bacteria Nachweis Von Säureproduzierenden Bakterien Détection De Bactéries Produisant Des Acides
(19) TZZ ¥ _T (11) EP 2 443 249 B1 (12) EUROPEAN PATENT SPECIFICATION (45) Date of publication and mention (51) Int Cl.: of the grant of the patent: C12Q 1/04 (2006.01) G01N 33/84 (2006.01) 19.11.2014 Bulletin 2014/47 (86) International application number: (21) Application number: 10790013.6 PCT/US2010/038569 (22) Date of filing: 15.06.2010 (87) International publication number: WO 2010/147918 (23.12.2010 Gazette 2010/51) (54) DETECTION OF ACID-PRODUCING BACTERIA NACHWEIS VON SÄUREPRODUZIERENDEN BAKTERIEN DÉTECTION DE BACTÉRIES PRODUISANT DES ACIDES (84) Designated Contracting States: (74) Representative: Isarpatent AL AT BE BG CH CY CZ DE DK EE ES FI FR GB Patent- und Rechtsanwälte GR HR HU IE IS IT LI LT LU LV MC MK MT NL NO Friedrichstrasse 31 PL PT RO SE SI SK SM TR 80801 München (DE) (30) Priority: 15.06.2009 US 187107 P (56) References cited: 15.03.2010 US 314140 P US-A- 4 528 269 US-A- 5 098 832 US-A- 5 164 301 US-A- 5 601 998 (43) Date of publication of application: US-A- 5 601 998 US-A- 5 786 167 25.04.2012 Bulletin 2012/17 US-B2- 6 756 225 US-B2- 7 150 977 (73) Proprietor: 3M Innovative Properties Company • DARUKARADHYA J ET AL: "Selective Saint Paul, MN 55133-3427 (US) enumeration of Lactobacillus acidophilus, Bifidobacterium spp., starter lactic acid bacteria (72) Inventors: and non-starter lactic acid bacteria from Cheddar • YOUNG, Robert, F. cheese", INTERNATIONAL DAIRY JOURNAL, Saint Paul, Minnesota 55133-3427 (US) ELSEVIER APPLIED SCIENCE, BARKING, GB, • MACH, Patrick, A. -

General Microbiology (11:680:390) Syllabus
COURSE SYLLABUS General Microbiology - 11:680:390 COURSE OVERVIEW General Microbiology 11:680:390 Fall, Spring, Summer Meeting times TBD Meeting Location Lecture: Sychronous Lecture Hall Cook/Douglas and Wright Labs Busch Meeting Location Lab: Food Science 209 CONTACT INFORMATION: Course Coordinator: Dr. Ines Rauschenbach Office Location: Lipman Hall, Room 215 Phone: 848-932-5635 Email: [email protected] Office Hours: By Appointment COURSE WEBSITE, RESOURCES AND MATERIALS: • Canvas • Text: Madigan MT, Bender KS, Buckley DH, Sattley WM, Stahl DA. 2020. Brock Biology of Microorganisms. 16th edition. Pearson, New York, NY. • Lab Manual o The lab manual (departmental publication) will be available for free through RUCore. • Electronic Notebook o We will be sending you a link to LabArchives. You must sign up before the start of your first lab. COURSE DESCRIPTION: This course offers a comprehensive study of the field of microbiology to science majors. The course will give detailed insights into five major themes: Structure and function of microbes (cellular structures, metabolism, and growth);,microbial genetics, microbial ecology, microbial diversity (prokaryotes, eukaryotes, viruses) and clinical microbiology (immunity, pathogenicity, epidemiology, control of microbes, and diseases). The course is taught in the synchronous lecture halls on Cook/Douglass and Busch campuses. Students are expected to participate in active learning activities and participate in class discussion to deepen their understanding of the microbial world and apply their knowledge to various concepts. LEARNING GOALS: Learning Goals for General Microbiology Lecture: After completion of the lecture component of the course, successful students will: 1. Demonstrate an understanding of the structural similarities and differences among microbes and the unique structure/function relationships of prokaryotic cells. -

Chemostat Culture for Yeast Experimental Evolution
Downloaded from http://cshprotocols.cshlp.org/ at Cold Spring Harbor Laboratory Library on August 9, 2017 - Published by Cold Spring Harbor Laboratory Press Protocol Chemostat Culture for Yeast Experimental Evolution Celia Payen and Maitreya J. Dunham1 Department of Genome Sciences, University of Washington, Seattle, Washington 98195 Experimental evolution is one approach used to address a broad range of questions related to evolution and adaptation to strong selection pressures. Experimental evolution of diverse microbial and viral systems has routinely been used to study new traits and behaviors and also to dissect mechanisms of rapid evolution. This protocol describes the practical aspects of experimental evolution with yeast grown in chemostats, including the setup of the experiment and sampling methods as well as best laboratory and record-keeping practices. MATERIALS It is essential that you consult the appropriate Material Safety Data Sheets and your institution’s Environmental Health and Safety Office for proper handling of equipment and hazardous material used in this protocol. Reagents Defined minimal medium appropriate for the experiment For examples, see Protocol: Assembly of a Mini-Chemostat Array (Miller et al. 2015). Ethanol (95%) Glycerol (20% and 50%; sterile) Yeast strain of interest Equipment Agar plates (appropriate for chosen strain) Chemostat array Assemble the apparatus as described in Miller et al. (2013) and Protocol: Assembly of a Mini-Chemostat Array (Miller et al. 2015). Cryo deep-freeze labels Cryogenic vials Culture tubes Cytometer (BD Accuri C6) Glass beads, 4 mm (sterile; for plating yeast cells) Glass cylinder Kimwipes 1Correspondence: [email protected] © 2017 Cold Spring Harbor Laboratory Press Cite this protocol as Cold Spring Harb Protoc; doi:10.1101/pdb.prot089011 559 Downloaded from http://cshprotocols.cshlp.org/ at Cold Spring Harbor Laboratory Library on August 9, 2017 - Published by Cold Spring Harbor Laboratory Press C. -

Microbiology Laboratory Exercises Third Edition 2020
MICROBIOLOGY Laboratory Exercises Third Edition Keddis & Rauschenbach 2020 Photo Credits (in order of contribution): Diane Davis, Ines Rauschenbach & Ramaydalis Keddis Acknowledgements: Many thanks to those in the Department of Biochemistry and Microbiology, Rutgers University, who have through the years inspired our enthusiasm for the science and teaching of microbiology, with special thanks to Diane Davis, Douglas Eveleigh and Max Häggblom. Safety: The experiments included in this manual have been deemed safe by the authors when all necessary safety precautions are met. The authors recommend maintaining biosafety level 2 in the laboratory setting and using risk level 1 organisms for all exercises. License: This work is licensed under a Creative Commons Attribution- NonCommercial-NoDerivatives 4.0 International License Microbiology Laboratory Exercises Third Edition 2020 Ramaydalis Keddis, Ph.D. Ines Rauschenbach, Ph.D. Department of Biochemistry and Microbiology Rutgers, The State University of New Jersey CONTENTS PAGE Introduction Schedule ii Best Laboratory Practices Iii Working in a Microbiology Laboratory iv Exercises Preparation of a Culture Medium 1 Culturing and Handling Microorganisms 3 Isolation of a Pure Culture 5 Counting Bacterial Populations 8 Controlling Microorganisms 10 Disinfectants 10 Antimicrobial Agents: Susceptibility Testing 12 Hand Washing 14 The Lethal Effects of Ultraviolet Light 15 Selection of Fungi from Air 17 Microscopy 21 Morphology and Staining of Bacteria 26 Microbial Metabolism 30 Enzyme Assay 32 Metabolic -
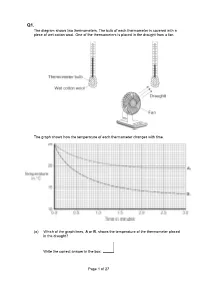
Page 1 of 27 the Diagram Shows Two Thermometers. the Bulb of Each
Q1. The diagram shows two thermometers. The bulb of each thermometer is covered with a piece of wet cotton wool. One of the thermometers is placed in the draught from a fan. The graph shows how the temperature of each thermometer changes with time. (a) Which of the graph lines, A or B, shows the temperature of the thermometer placed in the draught? Write the correct answer in the box. Page 1 of 27 Explain, in terms of evaporation, the reason for your answer. ___________________________________________________________________ ___________________________________________________________________ ___________________________________________________________________ ___________________________________________________________________ ___________________________________________________________________ ___________________________________________________________________ (3) (b) A wet towel spread out and hung outside on a day without wind dries faster than an identical wet towel left rolled up in a plastic bag. Explain why. ___________________________________________________________________ ___________________________________________________________________ ___________________________________________________________________ ___________________________________________________________________ (2) (Total 5 marks) Q2. The picture shows a person taking a hot shower. (a) When a person uses the shower the mirror gets misty. Why? ___________________________________________________________________ ___________________________________________________________________ -

Examination of Zoonotic Bacterial Pathogens in Canine and Feline Raw Meat-Based Diets
F.P.J. van Bree, Utrecht University, 2016 Paper Paper Examination of zoonotic bacterial pathogens in canine and feline raw meat-based diets F.P.J. van Bree, P.A.M. Overgaauw The feeding of raw meat-based diets (RMBD) to companion animals has become increasingly popular in recent years. However, such diets were demonstrated to be possibly contaminated with various zoonotic bacterial species in several studies and so the feeding of these diets could pose a risk to both animal and human health. Data about the situation in the Netherlands, or Europe in general are not available. Therefore, the purpose of this study was to evaluate the contamination of commercial RMBDs available in the Netherlands with zoonotic bacterial pathogens. Thirty-five commercial RMBDs were evaluated via bacterial culture for Escherichia coli O157:H7, extended- spectrum beta-lactamase (ESBL-)producing E. coli, Listeria monocytogenes and Salmonella spp.. E. coli O157:H7 was isolated from eight (23%) products, ESBL-producing E. coli from twenty- eight (80%) products, L. monocytogenes from nineteen (54%) products, other Listeria spp. from fifteen (43%) products, and Salmonella spp. from seven (20%) products. The results of this study demonstrate the need to pay attention to the potential of RMBDs to be a source for zoonotic bacterial pathogens in the Netherlands. These diets cannot be ruled out as a possible source for bacterial infections in both humans and animals and therefore companion animal owners should be informed about the associated risks. Keywords: raw meat-based diet; BARF; E. coli O157; ESBL; Listeria monocytogenes; Salmonella; public health In recent years it has become increasingly popular among of them are intended to provide for a complete diet for dogs dog and cat owners to feed raw meat-based diets (RMBD), and/or cats. -

Chemical Hygiene and Lab Safety Manual
CHEMICAL HYGIENE AND LAB SAFETY MANUAL SOUTH DAKOTA SCHOOL OF MINES & TECHNOLOGY Created: 01/12/05 Revision Date: 11/29/2018 FOR OFFICIAL USE ONLY Table of Contents CHEMICAL HYGIENE AND LAB SAFETY MANUAL ...................................................................................................................... 1 1. PURPOSE ...................................................................................................................................................................... 4 2. SCOPE ............................................................................................................................................................................ 4 3. DEFINITIONS (As excerpted from 29 CFR 1910.1450) .......................................................................................... 4 4. EMPLOYEE RIGHTS and RESPONSIBILITIES ....................................................................................................... 6 4.1 EMPLOYEE RIGHTS .......................................................................................................................................... 6 4.1 EMPLOYEE RESPONSIBILITIES ..................................................................................................................... 6 5. ENFORCEMENT ........................................................................................................................................................... 7 6. MEDICAL PROGRAM ................................................................................................................................................. -

Journal of Microbiology, Immunology and Infection
JOURNAL OF MICROBIOLOGY, IMMUNOLOGY AND INFECTION AUTHOR INFORMATION PACK TABLE OF CONTENTS XXX . • Description p.1 • Impact Factor p.1 • Abstracting and Indexing p.2 • Editorial Board p.2 • Guide for Authors p.4 ISSN: 1684-1182 DESCRIPTION . Journal of Microbiology, Immunology and Infection, launched in 1968, is the official bi-monthly publication of the Taiwan Society of Microbiology, the Chinese Society of Immunology, the Infectious Diseases Society of Taiwan and the Taiwan Society of Parasitology. The journal is an open access journal, committed to disseminating information on the latest trends and advances in microbiology, immunology, infectious diseases and parasitology. Articles on clinical or laboratory investigations of relevance to microbiology, immunology, infectious diseases, parasitology and other related fields that are of interest to the medical profession are eligible for consideration. Article types considered include perspectives, review articles, original articles, brief reports and correspondence. The Editorial Board of the Journal comprises a dedicated team of local and international experts in the field of microbiology, immunology, infectious diseases and parasitology. All members of the Editorial Board actively guide and set the direction of the journal. With the aim of promoting effective and accurate scientific information, an expert panel of referees constitutes the backbone of the peer- review process in evaluating the quality and content of manuscripts submitted for publication. JMII is open access and indexed in SCIE, PubMed, MEDLINE, EMBASE, Scopus, AIDS & Cancer Reseach, CABI, BIOSIS Previews, Biological Abstracts, EBSCOhost, CancerLit, Reactions Weekly (online), Chemical Abstracts, HealthSTAR, Global Health, ProQuest. Benefits to authors We also provide many author benefits, such as free PDFs, a liberal copyright policy, special discounts on Elsevier publications and much more. -

Performance Validation of the Microbiologique Microfilm Test
MAI ET AL.: JOURNAL OF AOAC INTERNATIONAL VOL. 101, NO. X, 2018 1 FOOD BIOLOGICAL CONTAMINANTS Performance Validation of the Microbiologique MicrofilmTM Test System for AOAC Research Institute Performance Tested Method SMCertification AOAC Performance Tested Method SM 051702 Abstract ANNA SHAPOVALOVA and HARISH K. JANAGAMA Molecular Epidemiology, Inc., 15300 Bothell Way NE, The Microfilm™ Test System is intended for quantitative Lake Forest Park, WA 98155 microbiology and consists of three types of Microfilms for aerobic LONG VUONG and ALEX FRIEDRICH plate count (Microfilm APC), total coliform and Escherichia coli IEH Laboratories and Consulting Group, Inc., 15300 Bothell count (Microfilm TCEc), and yeast and mold count (Microfilm Way NE, Lake Forest Park, WA 98155 YMC). This study evaluated the performance of the Microfilm DYLAN JOHNSON Test System against International Organization for Standardization Molecular Epidemiology, Inc., 15300 Bothell Way NE, (ISO) methods on 20 food matrixes and 2 environmental surfaces. Lake Forest Park, WA 98155 Ruggedness, robustness, and stability were also determined, LOURDES M. NADALA while inclusivity and exclusivity studies were performed on Molecular Epidemiology, Inc., 15300 Bothell Way N.E., Microfilm TCEc and YMC. An independent laboratory evaluated Lake Forest Park, WA 98155; Microbiologique, the performance on four food matrixes and one environmental Inc., 8215 Lake City Way NE, Seattle, WA 98115 surface. No significant differences and high correlation coefficients VAN NGUYEN were observed between the Microfilm Test System and the Microbiologique, Inc., 8215 Lake City Way NE, Seattle, corresponding ISO methods (ISO 4833-1:2013 for APC, ISO WA 98115 4832:2006 for total coliform count, ISO 16649-2: 2001 for E. -

Laboratory Exercises in Microbiology: Discovering the Unseen World Through Hands-On Investigation
City University of New York (CUNY) CUNY Academic Works Open Educational Resources Queensborough Community College 2016 Laboratory Exercises in Microbiology: Discovering the Unseen World Through Hands-On Investigation Joan Petersen CUNY Queensborough Community College Susan McLaughlin CUNY Queensborough Community College How does access to this work benefit ou?y Let us know! More information about this work at: https://academicworks.cuny.edu/qb_oers/16 Discover additional works at: https://academicworks.cuny.edu This work is made publicly available by the City University of New York (CUNY). Contact: [email protected] Laboratory Exercises in Microbiology: Discovering the Unseen World through Hands-On Investigation By Dr. Susan McLaughlin & Dr. Joan Petersen Queensborough Community College Laboratory Exercises in Microbiology: Discovering the Unseen World through Hands-On Investigation Table of Contents Preface………………………………………………………………………………………i Acknowledgments…………………………………………………………………………..ii Microbiology Lab Safety Instructions…………………………………………………...... iii Lab 1. Introduction to Microscopy and Diversity of Cell Types……………………......... 1 Lab 2. Introduction to Aseptic Techniques and Growth Media………………………...... 19 Lab 3. Preparation of Bacterial Smears and Introduction to Staining…………………...... 37 Lab 4. Acid fast and Endospore Staining……………………………………………......... 49 Lab 5. Metabolic Activities of Bacteria…………………………………………….…....... 59 Lab 6. Dichotomous Keys……………………………………………………………......... 77 Lab 7. The Effect of Physical Factors on Microbial Growth……………………………... 85 Lab 8. Chemical Control of Microbial Growth—Disinfectants and Antibiotics…………. 99 Lab 9. The Microbiology of Milk and Food………………………………………………. 111 Lab 10. The Eukaryotes………………………………………………………………........ 123 Lab 11. Clinical Microbiology I; Anaerobic pathogens; Vectors of Infectious Disease….. 141 Lab 12. Clinical Microbiology II—Immunology and the Biolog System………………… 153 Lab 13. Putting it all Together: Case Studies in Microbiology…………………………… 163 Appendix I.