Cerebellar Hypoplasia Information Sheet 06-7-19
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

TSEN54 Rabbit Pab Antibody
TSEN54 rabbit pAb antibody Catalog No : Source: Concentration : Mol.Wt. (Da): A22945 Rabbit 1 mg/ml 58819 Applications WB,IHC,ELISA Reactivity Human Dilution WB: 1:500 - 1:2000. IHC: 1:100 - 1:300. ELISA: 1:20000. Not yet tested in other applications. Storage -20°C/1 year Specificity TSEN54 Polyclonal Antibody detects endogenous levels of TSEN54 protein. Source / Purification The antibody was affinity-purified from rabbit antiserum by affinity- chromatography using epitope-specific immunogen. Immunogen The antiserum was produced against synthesized peptide derived from human TSEN54. AA range:261-310 Uniprot No Q7Z6J9 Alternative names TSEN54; SEN54; tRNA-splicing endonuclease subunit Sen54; SEN54 homolog; HsSEN54; tRNA-intron endonuclease Sen54 Form Liquid in PBS containing 50% glycerol, 0.5% BSA and 0.02% sodium azide. Clonality Polyclonal Isotype IgG Conjugation Background tRNA splicing endonuclease subunit 54(TSEN54) Homo sapiens This gene encodes a subunit of the tRNA splicing endonuclease complex, which catalyzes the removal of introns from precursor tRNAs. The complex is also implicated in pre-mRNA 3-prime end processing. Mutations in this gene result in pontocerebellar hypoplasia type 2.[provided by RefSeq, Oct 2009], Other TSEN54, tRNA-splicing endonuclease subunit Sen54 Produtc Images: Application Key: WB-Western IP-Immunoprecipitation IHC-Immunohistochemistry ChIP-Chromatin Immunoprecipitation A AAB Biosciences Products www.aabsci.cn FOR RESEARCH USE ONLY. NOT FOR HUMAN OR DIAGNOSTIC USE. IF-Immunofluorescence F-Flow Cytometry E-P-ELISA-Peptide Species Cross-Reactivity Key: H-Human M-Mouse R-Rat Hm-Hamster Mk-Monkey Vir-Virus Mi-Mink C-Chicken Dm-D. melanogaster X-Xenopus Z-Zebrafish B-Bovine Dg-Dog Pg-Pig Sc-S. -

Inherited Cerebellar Ataxia in Childhood: a Pattern-Recognition Approach Using Brain MRI
REVIEW ARTICLE Inherited Cerebellar Ataxia in Childhood: A Pattern-Recognition Approach Using Brain MRI L. Vedolin, G. Gonzalez, C.F. Souza, C. Lourenc¸o, and A.J. Barkovich ABSTRACT SUMMARY: Ataxia is the principal symptom of many common neurologic diseases in childhood. Ataxias caused by dysfunction of the cerebellum occur in acute, intermittent, and progressive disorders. Most of the chronic progressive processes are secondary to degen- erative and metabolic diseases. In addition, congenital malformation of the midbrain and hindbrain can also be present, with posterior fossa symptoms related to ataxia. Brain MR imaging is the most accurate imaging technique to investigate these patients, and imaging abnormalities include size, shape, and/or signal of the brain stem and/or cerebellum. Supratentorial and cord lesions are also common. This review will discuss a pattern-recognition approach to inherited cerebellar ataxia in childhood. The purpose is to provide a comprehensive discussion that ultimately could help neuroradiologists better manage this important topic in pediatric neurology. ABBREVIATIONS: AR ϭ autosomal recessive; CAC ϭ cerebellar ataxia in childhood; 4H ϭ hypomyelination with hypogonadotropic hypogonadism and hypodon- tia; JSRD ϭ Joubert syndrome and related disorders; OPHN1 ϭ oligophrenin-1 taxia is an inability to coordinate voluntary muscle move- plastic/paraneoplastic disorders, immune-mediated/demyelinat- Aments that cannot be attributed to weakness or involuntary ing disorders, and drugs/toxins (antiepileptic medications, -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -
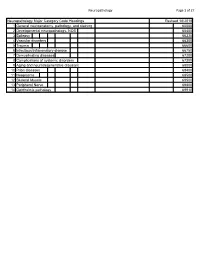
Neuropathology Category Code List
Neuropathology Page 1 of 27 Neuropathology Major Category Code Headings Revised 10/2018 1 General neuroanatomy, pathology, and staining 65000 2 Developmental neuropathology, NOS 65400 3 Epilepsy 66230 4 Vascular disorders 66300 5 Trauma 66600 6 Infectious/inflammatory disease 66750 7 Demyelinating diseases 67200 8 Complications of systemic disorders 67300 9 Aging and neurodegenerative diseases 68000 10 Prion diseases 68400 11 Neoplasms 68500 12 Skeletal Muscle 69500 13 Peripheral Nerve 69800 14 Ophthalmic pathology 69910 Neuropathology Page 2 of 27 Neuropathology 1 General neuroanatomy, pathology, and staining 65000 A Neuroanatomy, NOS 65010 1 Neocortex 65011 2 White matter 65012 3 Entorhinal cortex/hippocampus 65013 4 Deep (basal) nuclei 65014 5 Brain stem 65015 6 Cerebellum 65016 7 Spinal cord 65017 8 Pituitary 65018 9 Pineal 65019 10 Tracts 65020 11 Vascular supply 65021 12 Notochord 65022 B Cell types 65030 1 Neurons 65031 2 Astrocytes 65032 3 Oligodendroglia 65033 4 Ependyma 65034 5 Microglia and mononuclear cells 65035 6 Choroid plexus 65036 7 Meninges 65037 8 Blood vessels 65038 C Cerebrospinal fluid 65045 D Pathologic responses in neurons and axons 65050 1 Axonal degeneration/spheroid/reaction 65051 2 Central chromatolysis 65052 3 Tract degeneration 65053 4 Swollen/ballooned neurons 65054 5 Trans-synaptic neuronal degeneration 65055 6 Olivary hypertrophy 65056 7 Acute ischemic (hypoxic) cell change 65057 8 Apoptosis 65058 9 Protein aggregation 65059 10 Protein degradation/ubiquitin pathway 65060 E Neuronal nuclear inclusions 65100 -

Aneuploidy: Using Genetic Instability to Preserve a Haploid Genome?
Health Science Campus FINAL APPROVAL OF DISSERTATION Doctor of Philosophy in Biomedical Science (Cancer Biology) Aneuploidy: Using genetic instability to preserve a haploid genome? Submitted by: Ramona Ramdath In partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical Science Examination Committee Signature/Date Major Advisor: David Allison, M.D., Ph.D. Academic James Trempe, Ph.D. Advisory Committee: David Giovanucci, Ph.D. Randall Ruch, Ph.D. Ronald Mellgren, Ph.D. Senior Associate Dean College of Graduate Studies Michael S. Bisesi, Ph.D. Date of Defense: April 10, 2009 Aneuploidy: Using genetic instability to preserve a haploid genome? Ramona Ramdath University of Toledo, Health Science Campus 2009 Dedication I dedicate this dissertation to my grandfather who died of lung cancer two years ago, but who always instilled in us the value and importance of education. And to my mom and sister, both of whom have been pillars of support and stimulating conversations. To my sister, Rehanna, especially- I hope this inspires you to achieve all that you want to in life, academically and otherwise. ii Acknowledgements As we go through these academic journeys, there are so many along the way that make an impact not only on our work, but on our lives as well, and I would like to say a heartfelt thank you to all of those people: My Committee members- Dr. James Trempe, Dr. David Giovanucchi, Dr. Ronald Mellgren and Dr. Randall Ruch for their guidance, suggestions, support and confidence in me. My major advisor- Dr. David Allison, for his constructive criticism and positive reinforcement. -

GENETIC TESTING REQUISITION Please Ship All
GENETIC TESTING REQUISITION 1-844-363-4357· [email protected] Schillingallee 68 · 18057 Rostock Germany Attention Patient: Please visit your nearest LifeLabs or CML Healthcare Patient Service Centre for sample collection LL: K012-01/ CML: CEN CONTRACT # Report to Physician Billing # LifeLabs Demographic Ordering Physician Name Label Physician Signature: Ordering Physician Address: Tel: Fax: Address & Contact Info: Copy to (name & contact info): Name: Contact: Bill to Contract # K012-01 (patient does not pay at time of collection) Patient Gender: (M/F) Patient Name (Last, First): Patient DOB: (YYYY/MM/DD) Patient Address: Patient Health Card: Patient Telephone: Please ship all NON-PRENATAL samples to: LifeLabs · Attn CDS Department • 100 International Boulevard• Toronto ON• M9W6J6 TEST REQUESTED LL TR # / CML TC# □ Genetic Test - Blood Sample 2 x 4mL EDTA 4005 □ Genetic Test (Pediatric) - Blood Sample 1 x 2mL EDTA 4008 □ Genetic Test - Other Sample Type 4014 PRENATAL SAMPLES: Please ship directly to CENTOGENE. Date Blood Collected (YYYY/MM/DD): ___________ Time Blood Collected (HH:MM)) :________ Collector Name: ___________________ GENETIC TESTING CONSENT I understand that a DNA specimen will be sent to LifeLabs for genetic testing. My physician has told me about the condition(s) being tested and its genetic basis. I am aware that correct information about the relationships between my family members is important. I agree that my specimen and personal health information may be sent to Centogene AG at their lab in Germany (address below). To ensure accurate testing, I agree that the results of any genetic testing that I have had previously completed by Centogene AG may be shared with LifeLabs. -

NC RNA Symposium 2019 Fin
Organizing Committee: Co-Chairs of the Organizing Committee: Stacy Horner, Ph.D. (Duke) Robin Stanley, Ph.D. (NIEHS) Members of Organizing Committee: Christopher Holley, M.D, Ph.D. (Duke) Kate Meyer, Ph.D. (Duke) Hashim Al-Hashimi, Ph.D. (Duke) Hala Abou Assi, Ph.D. (Duke) Margot Wuebbens, Ph.D. (Duke) Sponsors: Symposium on RNA Biology XIII: RNA Tool and Target 2019 Meeting Agenda Thursday, October 17th, 2019 Great Hall of the Trent Semans Center, Duke University 12:00 – 1:00 PM Registration (Great Hall) 1:00 – 2:35 PM SESSION I: TRANSLATION FIDELITY Chair: Amanda Hargrove, Duke University 1:00 PM Opening Remarks, Stacy Horner, Duke University 1:10 PM Keynote Lecture: Rachel Green, Johns Hopkins University “Colliding Ribosomes as an integrator of Cytoplasmic Stress Responses” 1:50 PM Yu-Hua Lo, NIEHS “Unraveling the mechanism of substrate processing by the AAA-ATPase Rix7” 2:05 PM Hani Zaher, Washington University “RNA damage, the ribosome and quality control” 2:35 PM Break 3:00 – 5:00 PM SESSION II: RNA PROCESSING Chair: Jimena Giudice, UNC Chapel Hill 3:00 PM Rui Zhao, University of Colorado “A unified mechanism for intron definition, exon definition, and back-splicing” 3:30 PM Adam Black, UNC Chapel Hill “Clathrin heavy chain splice forms differentially affect striated muscle physiology” 3:45 PM Nicholas Conrad, University of Texas Southwestern “Mechanisms regulating S-adenosylmethionine homeostasis through intron detention” 4:15 PM Marcos Morgan, NIEHS “Role of RNA uridylation in germ line differentiation” 4:45 PM Jianguo Huang, -

Whole Exome Sequencing Gene Package Intellectual Disability, Version 9.1, 31-1-2020
Whole Exome Sequencing Gene package Intellectual disability, version 9.1, 31-1-2020 Technical information DNA was enriched using Agilent SureSelect DNA + SureSelect OneSeq 300kb CNV Backbone + Human All Exon V7 capture and paired-end sequenced on the Illumina platform (outsourced). The aim is to obtain 10 Giga base pairs per exome with a mapped fraction of 0.99. The average coverage of the exome is ~50x. Duplicate and non-unique reads are excluded. Data are demultiplexed with bcl2fastq Conversion Software from Illumina. Reads are mapped to the genome using the BWA-MEM algorithm (reference: http://bio-bwa.sourceforge.net/). Variant detection is performed by the Genome Analysis Toolkit HaplotypeCaller (reference: http://www.broadinstitute.org/gatk/). The detected variants are filtered and annotated with Cartagenia software and classified with Alamut Visual. It is not excluded that pathogenic mutations are being missed using this technology. At this moment, there is not enough information about the sensitivity of this technique with respect to the detection of deletions and duplications of more than 5 nucleotides and of somatic mosaic mutations (all types of sequence changes). HGNC approved Phenotype description including OMIM phenotype ID(s) OMIM median depth % covered % covered % covered gene symbol gene ID >10x >20x >30x A2ML1 {Otitis media, susceptibility to}, 166760 610627 66 100 100 96 AARS1 Charcot-Marie-Tooth disease, axonal, type 2N, 613287 601065 63 100 97 90 Epileptic encephalopathy, early infantile, 29, 616339 AASS Hyperlysinemia, -

Psykisk Utviklingshemming Og Forsinket Utvikling
Psykisk utviklingshemming og forsinket utvikling Genpanel, versjon v03 Tabellen er sortert på gennavn (HGNC gensymbol) Navn på gen er iht. HGNC >x10 Andel av genet som har blitt lest med tilfredstillende kvalitet flere enn 10 ganger under sekvensering x10 er forventet dekning; faktisk dekning vil variere. Gen Gen (HGNC Transkript >10x Fenotype (symbol) ID) AAAS 13666 NM_015665.5 100% Achalasia-addisonianism-alacrimia syndrome OMIM AARS 20 NM_001605.2 100% Charcot-Marie-Tooth disease, axonal, type 2N OMIM Epileptic encephalopathy, early infantile, 29 OMIM AASS 17366 NM_005763.3 100% Hyperlysinemia OMIM Saccharopinuria OMIM ABCB11 42 NM_003742.2 100% Cholestasis, benign recurrent intrahepatic, 2 OMIM Cholestasis, progressive familial intrahepatic 2 OMIM ABCB7 48 NM_004299.5 100% Anemia, sideroblastic, with ataxia OMIM ABCC6 57 NM_001171.5 93% Arterial calcification, generalized, of infancy, 2 OMIM Pseudoxanthoma elasticum OMIM Pseudoxanthoma elasticum, forme fruste OMIM ABCC9 60 NM_005691.3 100% Hypertrichotic osteochondrodysplasia OMIM ABCD1 61 NM_000033.3 77% Adrenoleukodystrophy OMIM Adrenomyeloneuropathy, adult OMIM ABCD4 68 NM_005050.3 100% Methylmalonic aciduria and homocystinuria, cblJ type OMIM ABHD5 21396 NM_016006.4 100% Chanarin-Dorfman syndrome OMIM ACAD9 21497 NM_014049.4 99% Mitochondrial complex I deficiency due to ACAD9 deficiency OMIM ACADM 89 NM_000016.5 100% Acyl-CoA dehydrogenase, medium chain, deficiency of OMIM ACADS 90 NM_000017.3 100% Acyl-CoA dehydrogenase, short-chain, deficiency of OMIM ACADVL 92 NM_000018.3 100% VLCAD -

Atypical Development of the Cerebellum : Impact on Language Function Lynette M
University of New Mexico UNM Digital Repository Psychology ETDs Electronic Theses and Dissertations 8-28-2012 Atypical development of the cerebellum : impact on language function Lynette M. Silva Follow this and additional works at: https://digitalrepository.unm.edu/psy_etds Recommended Citation Silva, Lynette M.. "Atypical development of the cerebellum : impact on language function." (2012). https://digitalrepository.unm.edu/psy_etds/129 This Dissertation is brought to you for free and open access by the Electronic Theses and Dissertations at UNM Digital Repository. It has been accepted for inclusion in Psychology ETDs by an authorized administrator of UNM Digital Repository. For more information, please contact [email protected]. Lynette M. Silva Candidate Psychology Department This dissertation is approved, and it is acceptable in quality and form for publication: Approved by the Dissertation Committee: Steven Verney, Co-Chairperson Ron Yeo, Co-Chairperson Robert Thoma Jean Lowe ATYPICAL DEVELOPMENT OF THE CEREBELLUM: IMPACT ON LANGUAGE FUNCTION By LYNETTE M. SILVA B.A., English, Stanford University, 1996 M.S., Psychology, University of New Mexico, 2009 DISSERTATION Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Psychology The University of New Mexico Albuquerque, New Mexico July 2012 iii Dedication For my parents, extended family, and valued friends, because it took a village. And for Martin Rodriguez, who served as my Virgil, and showed me the way. iv Acknowledgements I would like to thank Drs. Steven Verney, Ron Yeo, Robert Thoma, and Jean Lowe for their guidance and support as members of my dissertation committee. I am very thankful for the encouragement and supervision I continue to receive from Dr. -

New Tests 2 Test Updates 4 CPT Code Updates 11 Deleted Tests 11 Diagnostics Update Volume XXI, No
Volume XXI, No. 6 JUNE 2021 MONTHLY TEST UPDATES Diagnostics Update New Tests 2 Test Updates 4 CPT Code Updates 11 Deleted Tests 11 Diagnostics Update Volume XXI, No. 6 | JUNE 2021 New Tests Use Anti-DFS70 antibodies may help identify individuals who do not have an Anti-Carbamylated Protein (CarP) Antibody 520311 ANA-associated Autoimmune Rheumatic Disease (AARD) especially in the absence of significant clinical findings.1 Anti-DFS70 Ab, especially when positive CPT 83516 in isolation (i.e. in the absence of AARD-associated autoantibodies), may Synonyms Anti-CarP antigen antibody; RA marker prevent unnecessary referrals and examinations of ANA-positive individuals.2 Special Instructions This test has not been approved for NY state clients. Limitations This test should be used with clinical findings and other Specimen Serum autoimmune testing; it cannot be used alone to rule out autoimmune disease. Volume 1 mL This test was developed and its performance characteristics determined Minimum Volume 0.5 mL by Labcorp. It has not been cleared or approved by the Food and Drug Container Red-top tube; serum from red-top tube; serum from a gel tube; or Administration. serum gel tube Methodology Enzyme-linked immunosorbent assay (ELISA) Collection Separate serum from cells within one hour of collection. Transfer to a Additional Information Anti-DFS70 antibodies target the dense fine speckled plastic transport tube before shipping. protein of 70 kDa which is identical to Lens Epithelium-Derived Growth Factor Storage Instructions Refrigerate or freeze. or transcription co-activator p75 (LEDGFp75). They are detectable in 2% to 22% Stability of healthy individuals and in less than 1% of patients with AARD are of unknown Temperature Period clinical significance. -

Supplementary Table 1
Supplementary Table 1. 492 genes are unique to 0 h post-heat timepoint. The name, p-value, fold change, location and family of each gene are indicated. Genes were filtered for an absolute value log2 ration 1.5 and a significance value of p ≤ 0.05. Symbol p-value Log Gene Name Location Family Ratio ABCA13 1.87E-02 3.292 ATP-binding cassette, sub-family unknown transporter A (ABC1), member 13 ABCB1 1.93E-02 −1.819 ATP-binding cassette, sub-family Plasma transporter B (MDR/TAP), member 1 Membrane ABCC3 2.83E-02 2.016 ATP-binding cassette, sub-family Plasma transporter C (CFTR/MRP), member 3 Membrane ABHD6 7.79E-03 −2.717 abhydrolase domain containing 6 Cytoplasm enzyme ACAT1 4.10E-02 3.009 acetyl-CoA acetyltransferase 1 Cytoplasm enzyme ACBD4 2.66E-03 1.722 acyl-CoA binding domain unknown other containing 4 ACSL5 1.86E-02 −2.876 acyl-CoA synthetase long-chain Cytoplasm enzyme family member 5 ADAM23 3.33E-02 −3.008 ADAM metallopeptidase domain Plasma peptidase 23 Membrane ADAM29 5.58E-03 3.463 ADAM metallopeptidase domain Plasma peptidase 29 Membrane ADAMTS17 2.67E-04 3.051 ADAM metallopeptidase with Extracellular other thrombospondin type 1 motif, 17 Space ADCYAP1R1 1.20E-02 1.848 adenylate cyclase activating Plasma G-protein polypeptide 1 (pituitary) receptor Membrane coupled type I receptor ADH6 (includes 4.02E-02 −1.845 alcohol dehydrogenase 6 (class Cytoplasm enzyme EG:130) V) AHSA2 1.54E-04 −1.6 AHA1, activator of heat shock unknown other 90kDa protein ATPase homolog 2 (yeast) AK5 3.32E-02 1.658 adenylate kinase 5 Cytoplasm kinase AK7