Integrated Representation of Clinical Data and Medical Knowledge: An
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Sprouting Pinenut September 2018
1 Numa News Sprouting Pinenut September 2018 Fallon Paiute Shoshone Tribal Newsletter www.fpst.org Volume 12 Issue 9 September 2018 1 2 Laura Ijames Tribal Secretary Report [email protected] Our department participated in the Back to School Night which was held at the Fallon Convention Center. Jolene Thomas, Secretary’s Assistant, invested her time as a volunteer for the event where many backpacks and supplies given out for the new school year. It was good to see the whole community come out and support our youth. I would like to thank the FPST Co-Ed Softball team for donating their winning plaque to the tribe. Since the tribe donated to the team, they named the team after the tribe. I am glad to see that tribal donations are enriching our young adults in a positive way. If you would like to see the plaque and picture it will be in the lobby at the administration building. A successful antelope hunt The Labor Day weekend is coming soon and I hope everyone has a safe holiday weekend. Committee Openings: Land and Water Resources– 2 vacancies Senior Committee– 1 vacancies Election Committee Alternate-1vacancy Any interested Tribal Member may submit a Committee Appointment form to the FBC Secretary 3 1 4 5 1 6 7 1 8 FPST CO-ED SLOWPITCH CHAMPIONS Congratulations to the FPST Co-Ed Softball Team on their 1st place win in the Oasis Adult Softball Association League on Tuesday, August 8, 2018. The league winners received a 1st place plague as they showed why they are the top team in the league, with a big contin- gent of them tribal members. -

Gross Anatomy
www.BookOfLinks.com THE BIG PICTURE GROSS ANATOMY www.BookOfLinks.com Notice Medicine is an ever-changing science. As new research and clinical experience broaden our knowledge, changes in treatment and drug therapy are required. The authors and the publisher of this work have checked with sources believed to be reliable in their efforts to provide information that is complete and generally in accord with the standards accepted at the time of publication. However, in view of the possibility of human error or changes in medical sciences, neither the authors nor the publisher nor any other party who has been involved in the preparation or publication of this work warrants that the information contained herein is in every respect accurate or complete, and they disclaim all responsibility for any errors or omissions or for the results obtained from use of the information contained in this work. Readers are encouraged to confirm the infor- mation contained herein with other sources. For example and in particular, readers are advised to check the product information sheet included in the package of each drug they plan to administer to be certain that the information contained in this work is accurate and that changes have not been made in the recommended dose or in the contraindications for administration. This recommendation is of particular importance in connection with new or infrequently used drugs. www.BookOfLinks.com THE BIG PICTURE GROSS ANATOMY David A. Morton, PhD Associate Professor Anatomy Director Department of Neurobiology and Anatomy University of Utah School of Medicine Salt Lake City, Utah K. Bo Foreman, PhD, PT Assistant Professor Anatomy Director University of Utah College of Health Salt Lake City, Utah Kurt H. -

Vista Comparison to the Commercial Electronic Health Record Marketplace Final Report
VistA Comparison to the Commercial Electronic Health Record Marketplace Final Report February 4, 2011 Prepared for: U.S. Department of Veterans Affairs Martin Geffen Vice President Gartner Consulting [email protected] GARTNER CONSULTING This presentation, including any supporting materials, is owned by Gartner, Inc. and/or its affiliates and is for the sole use of the intended Gartner audience or other authorized recipients. This presentation may contain information that is confidential, proprietary or otherwise legally protected, and it may not be further copied, distributed or publicly displayed without the express written permission of Gartner, Inc. or its affiliates. © 2010 Gartner, Inc. and/or its affiliates. All rights reserved. Table of Contents ■ Executive Summary ■ Project Objectives and Approach ■ Electronic Health Record Systems Marketplace ■ VistA Overall Findings ■ VistA Capability Assessment ■ Appendices © 2011 Gartner, Inc. and/or its affiliates. All rights reserved. Gartner is a registered trademark of Gartner, Inc. or its affiliates. 1 Executive Summary © 2011 Gartner, Inc. and/or its affiliates. All rights reserved. Gartner is a registered trademark of Gartner, Inc. or its affiliates. 2 Objective and Approach ■ Project Objective – The Department of Veterans Affairs (VA) engaged Gartner to develop a fact-based assessment of how VistA capabilities compare to those that are found in leading commercial off-the-shelf (COTS) Electronic Health Record (EHR) products. ■ Approach – Gartner applied an evaluation framework which is based on Gartner research, Gartner‘s definition of an EHR and methodologies (e.g., Magic Quadrant, Generations Model, Hype Cycle) to compare VistA clinical functionality to that of the EHR COTS products. – The evaluation framework compared VistA capabilities to those of COTS products in three major categories: • Core Clinical Capabilities • Support for Key Care Venues • Support for select Stakeholders © 2011 Gartner, Inc. -

AC Group's 2007 Annual Report the Digital Medical Office of the Future
AC Group’s 2007 Annual Report The Digital Medical Office of the Future Computer Systems for the Physician’s Office May 2007 Comprehensive Report on: Overview of Physician adoption The Six Levels of Physician IT Maturity Practice Management Marketplace Secured Internet Document Image Management PDA and Mobile Healthcare Electronic Medical Record Functionality Electronic Medical Health Marketplace Regional Healthcare Information Organizations Mark R. Anderson, CPHIMS, FHIMSS Healthcare IT Futurist AC Group, Inc. (v) 281-413-5572 [email protected] www.acgroup.org AC Group’s 2007 Annual Report The Digital Medical Office of the Future Computer Systems for the Physician’s Office More about the Author Mr. Anderson is one of the nation's premier IT research futurists dedicated to health care. He is one of the leading national speakers on healthcare and physician practices and has spoken at more than 350 conferences and meetings since 2000. He has spent the last 30+ years focusing on Healthcare – not just technology questions, but strategic, policy, and organizational considerations. He tracks industry trends, conducts member surveys and case studies, assesses best practices, and performs benchmarking studies. Besides serving at the CEO of AC Group, Mr. Anderson served as the interim CIO for the Taconic IPA in 2004-05 (a 500 practice, 2,300+ physician IPA located in upper New York). Prior to joining AC Group, Inc. in February of 2000, Mr. Anderson was the worldwide head and VP of healthcare for META Group, Inc., the Chief Information Officer (CIO) with West Tennessee Healthcare, the Corporate CIO for the Sisters of Charity of Nazareth Health System, the Corporate Internal IT Consultant with the Sisters of Providence (SOP) Hospitals, and the Executive Director for Management Services for Denver Health and Hospitals and Harris County Hospital District. -

Human Anatomy As Related to Tumor Formation Book Four
SEER Program Self Instructional Manual for Cancer Registrars Human Anatomy as Related to Tumor Formation Book Four Second Edition U.S. DEPARTMENT OF HEALTH AND HUMAN SERVICES Public Health Service National Institutesof Health SEER PROGRAM SELF-INSTRUCTIONAL MANUAL FOR CANCER REGISTRARS Book 4 - Human Anatomy as Related to Tumor Formation Second Edition Prepared by: SEER Program Cancer Statistics Branch National Cancer Institute Editor in Chief: Evelyn M. Shambaugh, M.A., CTR Cancer Statistics Branch National Cancer Institute Assisted by Self-Instructional Manual Committee: Dr. Robert F. Ryan, Emeritus Professor of Surgery Tulane University School of Medicine New Orleans, Louisiana Mildred A. Weiss Los Angeles, California Mary A. Kruse Bethesda, Maryland Jean Cicero, ART, CTR Health Data Systems Professional Services Riverdale, Maryland Pat Kenny Medical Illustrator for Division of Research Services National Institutes of Health CONTENTS BOOK 4: HUMAN ANATOMY AS RELATED TO TUMOR FORMATION Page Section A--Objectives and Content of Book 4 ............................... 1 Section B--Terms Used to Indicate Body Location and Position .................. 5 Section C--The Integumentary System ..................................... 19 Section D--The Lymphatic System ....................................... 51 Section E--The Cardiovascular System ..................................... 97 Section F--The Respiratory System ....................................... 129 Section G--The Digestive System ......................................... 163 Section -

Collaborative Stage Manual Part II
SEER Program Coding and Staging Manual 2004, Revision 1 Collaborative Staging Codes Nasal Cavity C30.0 C30.0 Nasal cavity (excludes nose, NOS C76.0) Note: Laterality must be coded for this site, except subsites Nasal cartilage and Nasal septum, for which laterality is coded 0. CS Tumor Size CS Site-Specific Factor 1 - Size of The following tables are available CS Extension Lymph Nodes at the collaborative staging CS TS/Ext-Eval CS Site-Specific Factor 2 - website: CS Lymph Nodes Extracapsular Extension, Lymph Nodes Histology Exclusion Table CS Reg Nodes Eval for Head and Neck AJCC Stage Reg LN Pos CS Site-Specific Factor 3 - Levels I- Lymph Nodes Size Table Reg LN Exam III, Lymph Nodes for Head and Neck CS Mets at DX CS Site-Specific Factor 4 - Levels IV- CS Mets Eval V and Retropharyngeal Lymph Nodes for Head and Neck CS Site-Specific Factor 5 - Levels VI- VII and Facial Lymph Nodes for Head and Neck CS Site-Specific Factor 6 - Parapharyngeal, Parotid, Preauricular, and Sub-Occipital Lymph Nodes, Lymph Nodes for Head and Neck Nasal Cavity CS Tumor Size SEE STANDARD TABLE Nasal Cavity CS Extension Code Description TNM SS77 SS2000 00 In situ; non-invasive Tis IS IS 10 Invasive tumor confined to site of origin T1 L L Meatus (superior, middle, inferior) Nasal chonchae (superior, middle, inferior) Septum Tympanic membrane 30 Localized, NOS T1 L L 40 Extending to adjacent connective tissue within the nasoethomoidal T2 RE RE complex Nasolacrimal duct 60 Adjacent organs/structures including: T3 RE RE Bone of skull Choana Frontal sinus Hard palate -

Normal Gross and Histologic Features of the Gastrointestinal Tract
NORMAL GROSS AND HISTOLOGIC 1 FEATURES OF THE GASTROINTESTINAL TRACT THE NORMAL ESOPHAGUS left gastric, left phrenic, and left hepatic accessory arteries. Veins in the proximal and mid esopha- Anatomy gus drain into the systemic circulation, whereas Gross Anatomy. The adult esophagus is a the short gastric and left gastric veins of the muscular tube measuring approximately 25 cm portal system drain the distal esophagus. Linear and extending from the lower border of the cri- arrays of large caliber veins are unique to the distal coid cartilage to the gastroesophageal junction. esophagus and can be a helpful clue to the site of It lies posterior to the trachea and left atrium a biopsy when extensive cardiac-type mucosa is in the mediastinum but deviates slightly to the present near the gastroesophageal junction (4). left before descending to the diaphragm, where Lymphatic vessels are present in all layers of the it traverses the hiatus and enters the abdomen. esophagus. They drain to paratracheal and deep The subdiaphragmatic esophagus lies against cervical lymph nodes in the cervical esophagus, the posterior surface of the left hepatic lobe (1). bronchial and posterior mediastinal lymph nodes The International Classification of Diseases in the thoracic esophagus, and left gastric lymph and the American Joint Commission on Cancer nodes in the abdominal esophagus. divide the esophagus into upper, middle, and lower thirds, whereas endoscopists measure distance to points in the esophagus relative to the incisors (2). The esophagus begins 15 cm from the incisors and extends 40 cm from the incisors in the average adult (3). The upper and lower esophageal sphincters represent areas of increased resting tone but lack anatomic landmarks; they are located 15 to 18 cm from the incisors and slightly proximal to the gastroesophageal junction, respectively. -

Semi-Annual Market Review
Semi-Annual Market Review HEALTH IT & HEALTH INFORMATION SERVICES JULY 2019 www.hgp.com TABLE OF CONTENTS 1 Health IT Executive Summary 3 2 Health IT Market Trends 6 3 HIT M&A (Including Buyout) 9 4 Health IT Capital Raises (Non-Buyout) 14 5 Healthcare Capital Markets 15 6 Macroeconomics 19 7 Health IT Headlines 21 8 About Healthcare Growth Partners 24 9 HGP Transaction Experience 25 10 Appendix A – M&A Highlights 28 11 Appendix B – Buyout Highlights 31 12 Appendix C – Investment Highlights 34 Copyright© 2019 Healthcare Growth Partners 2 HEALTH IT EXECUTIVE SUMMARY 1 An Accumulating Backlog of Disciplined Sellers Let’s chat about fireside chats. The term first used to describe a series of evening radio addresses given by U.S. President Franklin D. Roosevelt during the Great Depression and World War II is now investment banker speak for “soft launches” of sell-side and capital raise transactions. Every company has a price, and given a market of healthy valuations, more companies are testing the waters to find out whether they can achieve that price. That process now looks a little more informal, or how you might envision a fireside chat. Price (or valuation) discovery for a company can range from a single conversation with an individual buyer to a full-blown auction with hundreds of buyers and everything in between, including a fireside chat. Given the increasing share of informal conversations, the reality is that more companies are for sale than meets the eye. While the healthy valuations publicized and press-released are encouraging more and more companies to price shop, there is a simultaneous statistical phenomenon in perceived valuations that often goes unmentioned: survivorship bias. -

Private Equity Buyouts in Healthcare: Who Wins, Who Loses? Eileen Appelbaum and Rosemary Batt Working Paper No
Private Equity Buyouts in Healthcare: Who Wins, Who Loses? Eileen Appelbaum* and Rosemary Batt† Working Paper No. 118 March 15, 2020 ABSTRACT Private equity firms have become major players in the healthcare industry. How has this happened and what are the results? What is private equity’s ‘value proposition’ to the industry and to the American people -- at a time when healthcare is under constant pressure to cut costs and prices? How can PE firms use their classic leveraged buyout model to ‘save healthcare’ while delivering ‘outsized returns’ to investors? In this paper, we bring together a wide range of sources and empirical evidence to answer these questions. Given the complexity of the sector, we focus on four segments where private equity firms have been particularly active: hospitals, outpatient care (urgent care and ambulatory surgery centers), physician staffing and emergency * Co-Director and Senior Economist, Center for Economic and Policy Research. [email protected] † Alice H. Cook Professor of Women and Work, HR Studies and Intl. & Comparative Labor ILR School, Cornell University. [email protected]. We thank Andrea Beaty, Aimee La France, and Kellie Franzblau for able research assistance. room services (surprise medical billing), and revenue cycle management (medical debt collecting). In each of these segments, private equity has taken the lead in consolidating small providers, loading them with debt, and rolling them up into large powerhouses with substantial market power before exiting with handsome returns. https://doi.org/10.36687/inetwp118 JEL Codes: I11 G23 G34 Keywords: Private Equity, Leveraged Buyouts, health care industry, financial engineering, surprise medical billing revenue cycle management, urgent care, ambulatory care. -

Ministry of Education and Science of Ukraine Sumy State University 0
Ministry of Education and Science of Ukraine Sumy State University 0 Ministry of Education and Science of Ukraine Sumy State University SPLANCHNOLOGY, CARDIOVASCULAR AND IMMUNE SYSTEMS STUDY GUIDE Recommended by the Academic Council of Sumy State University Sumy Sumy State University 2016 1 УДК 611.1/.6+612.1+612.017.1](072) ББК 28.863.5я73 С72 Composite authors: V. I. Bumeister, Doctor of Biological Sciences, Professor; L. G. Sulim, Senior Lecturer; O. O. Prykhodko, Candidate of Medical Sciences, Assistant; O. S. Yarmolenko, Candidate of Medical Sciences, Assistant Reviewers: I. L. Kolisnyk – Associate Professor Ph. D., Kharkiv National Medical University; M. V. Pogorelov – Doctor of Medical Sciences, Sumy State University Recommended for publication by Academic Council of Sumy State University as а study guide (minutes № 5 of 10.11.2016) Splanchnology Cardiovascular and Immune Systems : study guide / С72 V. I. Bumeister, L. G. Sulim, O. O. Prykhodko, O. S. Yarmolenko. – Sumy : Sumy State University, 2016. – 253 p. This manual is intended for the students of medical higher educational institutions of IV accreditation level who study Human Anatomy in the English language. Посібник рекомендований для студентів вищих медичних навчальних закладів IV рівня акредитації, які вивчають анатомію людини англійською мовою. УДК 611.1/.6+612.1+612.017.1](072) ББК 28.863.5я73 © Bumeister V. I., Sulim L G., Prykhodko О. O., Yarmolenko O. S., 2016 © Sumy State University, 2016 2 Hippocratic Oath «Ὄμνυμι Ἀπόλλωνα ἰητρὸν, καὶ Ἀσκληπιὸν, καὶ Ὑγείαν, καὶ Πανάκειαν, καὶ θεοὺς πάντας τε καὶ πάσας, ἵστορας ποιεύμενος, ἐπιτελέα ποιήσειν κατὰ δύναμιν καὶ κρίσιν ἐμὴν ὅρκον τόνδε καὶ ξυγγραφὴν τήνδε. -
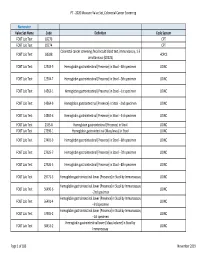
2020 Measure Value Set Colorectal Cancer Screening
PT ‐ 2020 Measure Value Set_Colorectal Cancer Screening Numerator Value Set Name Code Definition Code System FOBT Lab Test 82270 CPT FOBT Lab Test 82274 CPT Colorectal cancer screening; fecal occult blood test, immunoassay, 1‐3 FOBT Lab Test G0328 HCPCS simultaneous (G0328) FOBT Lab Test 12503‐9 Hemoglobin.gastrointestinal [Presence] in Stool ‐‐4th specimen LOINC FOBT Lab Test 12504‐7 Hemoglobin.gastrointestinal [Presence] in Stool ‐‐5th specimen LOINC FOBT Lab Test 14563‐1 Hemoglobin.gastrointestinal [Presence] in Stool ‐‐1st specimen LOINC FOBT Lab Test 14564‐9 Hemoglobin.gastrointestinal [Presence] in Stool ‐‐2nd specimen LOINC FOBT Lab Test 14565‐6 Hemoglobin.gastrointestinal [Presence] in Stool ‐‐3rd specimen LOINC FOBT Lab Test 2335‐8 Hemoglobin.gastrointestinal [Presence] in Stool LOINC FOBT Lab Test 27396‐1 Hemoglobin.gastrointestinal [Mass/mass] in Stool LOINC FOBT Lab Test 27401‐9 Hemoglobin.gastrointestinal [Presence] in Stool ‐‐6th specimen LOINC FOBT Lab Test 27925‐7 Hemoglobin.gastrointestinal [Presence] in Stool ‐‐7th specimen LOINC FOBT Lab Test 27926‐5 Hemoglobin.gastrointestinal [Presence] in Stool ‐‐8th specimen LOINC FOBT Lab Test 29771‐3 Hemoglobin.gastrointestinal.lower [Presence] in Stool by Immunoassay LOINC Hemoglobin.gastrointestinal.lower [Presence] in Stool by Immunoassay FOBT Lab Test 56490‐6 LOINC ‐‐2nd specimen Hemoglobin.gastrointestinal.lower [Presence] in Stool by Immunoassay FOBT Lab Test 56491‐4 LOINC ‐‐3rd specimen Hemoglobin.gastrointestinal.lower [Presence] in Stool by Immunoassay FOBT Lab Test 57905‐2 -

To See 2014 Health Policy Survey Results
2014 HEALTH POLICY SURVEY RESULTS Western Section American Urological Association Jeffrey M. Frankel, M.D. Chairman, Health Policy Committee An educational supplement to the WSAUA Health Policy Forum, October 26, 2014 – Grand Wailea Hotel, Maui, Hawaii 2014 Health Policy Committee Chairman: Jeffrey M. Frankel, M.D. Vice-Chairman: Eugene Rhee, M.D., MBA Robert G. Carlile, M.D., Hawaii District 1 Jeffrey M. Frankel, M.D., Washington District 2 Brian S. Shaffer, M.D., Portland District 3 Joseph R. Kuntze, M.D., Fresno District 4 Demetrios Simopoulas M.D., Sacramento District 5 Jeremy Shelton, M.D., Los Angeles District 6 Ithaar Derweesh, M.D., San Diego District 7 Micheal Darson, M.D., Scottsdale District 8 Lane Childs, M.D., Salt Lake City District 9 Aaron Spitz, M.D., Orange County District 10 Advisors: • Jeffrey E. Kaufman, MD • Amani Abou Zam-zam, MBA Copyright2014 by the Western Section American Urological Association, Inc. All rights reserved. Published by the Western Section American Urological Association, Inc. 1950 Old Tustin Avenue, Santa Ana, CA 92705. Call 714-550-9155 to order copies. Geographical Districts of the Western Section of the American Urological Association, Inc. District 1: Northwest Canadian Provinces, Alaska, Hawaii, Philippine Islands and Pacific Island Possessions of the U.S.A. District 2: Washington District 3: Oregon District 4: Northern California including Alameda and Contra Costa Counties District 5: San Francisco, San Mateo, and Santa Clara Counties and Central California District 6: Los Angeles County District