Molecular Cloning and Functional Characterization of a Dihydroflavonol 4-Reductase from Vitis Bellula
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Wo 2007/133479 A2
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (43) International Publication Date (10) International Publication Number 22 November 2007 (22.11.2007) PCT WO 2007/133479 A2 (51) International Patent Classification: Not classified (74) Agent: GODLEWSKI, Richard, J.; P.O. Box 2269, Bloomington, IN 47402-2269 (US). (21) International Application Number: (81) Designated States (unless otherwise indicated, for every PCT/US2007/0 10828 kind of national protection available): AE, AG, AL, AM, AT,AU, AZ, BA, BB, BG, BH, BR, BW, BY, BZ, CA, CH, (22) International Filing Date: 4 May 2007 (04.05.2007) CN, CO, CR, CU, CZ, DE, DK, DM, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, HR, HU, ID, IL, IN, (25) Filing Language: English IS, JP, KE, KG, KM, KN, KP, KR, KZ, LA, LC, LK, LR, LS, LT, LU, LY,MA, MD, ME, MG, MK, MN, MW, MX, (26) Publication Language: English MY, MZ, NA, NG, NI, NO, NZ, OM, PG, PH, PL, PT, RO, RS, RU, SC, SD, SE, SG, SK, SL, SM, SV, SY, TJ, TM, (30) Priority Data: TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW 60/799,608 10 May 2006 (10.05.2006) US (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, (71) Applicants (for all designated States except US): COOK GM, KE, LS, MW, MZ, NA, SD, SL, SZ, TZ, UG, ZM, INCORPORATED [US/US]; 750 North Daniel's Way, ZW), Eurasian (AM, AZ, BY, KG, KZ, MD, RU, TJ, TM), P.O. -

Sources of Crude Drug, Plant Families, Biogenesis of Phytochemicals
1 Sources of Crude Drug, Plant Families, Biogenesis of Phytochemicals SOURCES OF CRUDE DRUG Plant Oldest source of drugs. 25% of the drugs prescribed worldwide come from plants More than 200 drugs considered as basic and essential by the World Health Organisation (WHO) Significant number of synthetic drugs obtained from natural precursors. Example: Digoxin from Digitalis species, quinine and quinidine from Cinchona species, vincristrine and vinblastine from Catharanthus roseus, atropine from Atropa belladonna and morphine and codeine from Papaver somniferum. Animal Second largest source of crude drugs. Example: Honey from honeybee, beeswax from bees, cod liver oil from shark, bufalin from toad, animal pancreas is a source of Insulin, musk oil from musk, spermaceti wax from sperm whale, woolfat from sheep, carminic acid from colchineal, venoms from snake Mineral Highly purified form of naturally occurring mineral substances is used in medicine Example: Sulphur is a key ingredient in certain bacteriostatic drugs, shilajit is used as tonic, calamine is used as anti-itching agent Marine Major part of earth is covered with water bodies and hence bioactive compounds from marine flora and fauna (microorganisms, algae, fungi, invertebrates, and 1 Contd… 2 Pharmacognosy and Phytochemistry: A Companion Handbook vertebrates) have extensive past and present use in the treatment of many diseases Marine Serve as compounds of interest both in their natural form and as templates for synthetic modification. Several molecules isolated from various -

Phenolic Compounds in Cereal Grains and Their Health Benefits
and antioxidant activity are reported in the Phenolic Compounds in Cereal literature. Unfortunately, it is difficult to make comparisons of phenol and anti- Grains and Their Health Benefits oxidant activity levels in cereals since different methods have been used. The ➤ Whole grain cereals are a good source of phenolics. purpose of this article is to give an overview ➤ Black sorghums contain high levels of the unique 3-deoxyanthocyanidins. of phenolic compounds reported in whole ➤ Oats are the only source of avenanthramides. grain cereals and to compare their phenol and antioxidant activity levels. ➤ Among cereal grains, tannin sorghum and black rice contain the highest antioxidant activity in vitro. Phenolic Acids Phenolic acids are derivatives of benzoic and cinnamic acids (Fig. 1) and are present in all cereals (Table I). There are two Most of the literature on plant phenolics classes of phenolic acids: hydroxybenzoic L. DYKES AND L. W. ROONEY focuses mainly on those in fruits, acids and hydroxycinnamic acids. Hy- TEXAS A&M UNIVERSITY vegetables, wines, and teas (33,50,53,58, droxybenzoic acids include gallic, p- College Station, TX 74). However, many phenolic compounds hydroxybenzoic, vanillic, syringic, and in fruits and vegetables (i.e., phenolic acids protocatechuic acids. The hydroxycinna- esearch has shown that whole grain and flavonoids) are also reported in cereals. mic acids have a C6-C3 structure and Rconsumption helps lower the risk of The different species of grains have a great include coumaric, caffeic, ferulic, and cardiovascular disease, ischemic stroke, deal of diversity in their germplasm sinapic acids. The phenolic acids reported type II diabetes, metabolic syndrome, and resources, which can be exploited. -

The Science of Flavonoids the Science of Flavonoids
The Science of Flavonoids The Science of Flavonoids Edited by Erich Grotewold The Ohio State University Columbus, Ohio, USA Erich Grotewold Department of Cellular and Molecular Biology The Ohio State University Columbus, Ohio 43210 USA [email protected] The background of the cover corresponds to the accumulation of flavonols in the plasmodesmata of Arabidopsis root cells, as visualized with DBPA (provided by Dr. Wendy Peer). The structure corresponds to a model of the Arabidopsis F3 'H enzyme (provided by Dr. Brenda Winkel). The chemical structure corresponds to dihydrokaempferol. Library of Congress Control Number: 2005934296 ISBN-10: 0-387-28821-X ISBN-13: 978-0387-28821-5 ᭧2006 Springer ScienceϩBusiness Media, Inc. All rights reserved. This work may not be translated or copied in whole or in part without the written permission of the publisher (Springer ScienceϩBusiness Media, Inc., 233 Spring Street, New York, NY 10013, USA), except for brief excerpts in connection with reviews or scholarly analysis. Use in connection with any form of information storage and retrieval, electronic adaptation, computer software, or by similar or dissimilar methodology now known or hereafter developed is forbidden. The use in this publication of trade names, trademarks, service marks and similar terms, even if they are not identified as such, is not to be taken as an expression of opinion as to whether or not they are subject to proprietary rights. Printed in the United States of America (BS/DH) 987654321 springeronline.com PREFACE There is no doubt that among the large number of natural products of plant origin, debatably called secondary metabolites because their importance to the eco- physiology of the organisms that accumulate them was not initially recognized, flavonoids play a central role. -

Anthocyanin Pigments: Beyond Aesthetics
molecules Review Anthocyanin Pigments: Beyond Aesthetics , Bindhu Alappat * y and Jayaraj Alappat y Warde Academic Center, St. Xavier University, 3700 W 103rd St, Chicago, IL 60655, USA; [email protected] * Correspondence: [email protected] These authors contributed equally to this work. y Academic Editor: Pasquale Crupi Received: 29 September 2020; Accepted: 19 November 2020; Published: 24 November 2020 Abstract: Anthocyanins are polyphenol compounds that render various hues of pink, red, purple, and blue in flowers, vegetables, and fruits. Anthocyanins also play significant roles in plant propagation, ecophysiology, and plant defense mechanisms. Structurally, anthocyanins are anthocyanidins modified by sugars and acyl acids. Anthocyanin colors are susceptible to pH, light, temperatures, and metal ions. The stability of anthocyanins is controlled by various factors, including inter and intramolecular complexations. Chromatographic and spectrometric methods have been extensively used for the extraction, isolation, and identification of anthocyanins. Anthocyanins play a major role in the pharmaceutical; nutraceutical; and food coloring, flavoring, and preserving industries. Research in these areas has not satisfied the urge for natural and sustainable colors and supplemental products. The lability of anthocyanins under various formulated conditions is the primary reason for this delay. New gene editing technologies to modify anthocyanin structures in vivo and the structural modification of anthocyanin via semi-synthetic methods offer new opportunities in this area. This review focusses on the biogenetics of anthocyanins; their colors, structural modifications, and stability; their various applications in human health and welfare; and advances in the field. Keywords: anthocyanins; anthocyanidins; biogenetics; polyphenols; flavonoids; plant pigments; anthocyanin bioactivities 1. Introduction Anthocyanins are water soluble pigments that occur in most vascular plants. -

( 12 ) United States Patent
US010722444B2 (12 ) United States Patent ( 10 ) Patent No.: US 10,722,444 B2 Gousse et al. (45 ) Date of Patent : Jul. 28 , 2020 (54 ) STABLE HYDROGEL COMPOSITIONS 4,605,691 A 8/1986 Balazs et al . 4,636,524 A 1/1987 Balazs et al . INCLUDING ADDITIVES 4,642,117 A 2/1987 Nguyen et al. 4,657,553 A 4/1987 Taylor (71 ) Applicant: Allergan Industrie , SAS , Pringy (FR ) 4,713,448 A 12/1987 Balazs et al . 4,716,154 A 12/1987 Malson et al. ( 72 ) 4,772,419 A 9/1988 Malson et al. Inventors: Cécile Gousse , Dingy St. Clair ( FR ) ; 4,803,075 A 2/1989 Wallace et al . Sébastien Pierre, Annecy ( FR ) ; Pierre 4,886,787 A 12/1989 De Belder et al . F. Lebreton , Annecy ( FR ) 4,896,787 A 1/1990 Delamour et al. 5,009,013 A 4/1991 Wiklund ( 73 ) Assignee : Allergan Industrie , SAS , Pringy (FR ) 5,087,446 A 2/1992 Suzuki et al. 5,091,171 A 2/1992 Yu et al. 5,143,724 A 9/1992 Leshchiner ( * ) Notice : Subject to any disclaimer , the term of this 5,246,698 A 9/1993 Leshchiner et al . patent is extended or adjusted under 35 5,314,874 A 5/1994 Miyata et al . U.S.C. 154 (b ) by 0 days. 5,328,955 A 7/1994 Rhee et al . 5,356,883 A 10/1994 Kuo et al . (21 ) Appl . No.: 15 /514,329 5,399,351 A 3/1995 Leshchiner et al . 5,428,024 A 6/1995 Chu et al . -

Bbm:978-3-540-73733-9/1.Pdf
Sachverzeichnis Abelmoschus esculenta 481 Aflatoxine 449 Abelmoschus esculentus 491 Afzelechin 469 Abietinsäure 26 Agar-Agar 362, 402, 412 Acacetin 478 Agarose 412, 413 Acacia 402 Agave sisalana 69, 70 Acanthoscelides obtectus 648, 649 Aglycon 353 Acarbose 372, 399, 400, 401 Agmatin 151 ACE 292 Agnosterin 34 Acetoacetyl-ACP 575 AIDS 410 Acetobacter suboxidans 375 Ajmalicin 200 α-Acetobromglucose 355 Ajmalin 196 Acetoide 5, 43 Ajuga ripponesis 391 Acetylcholin 143ff, 148, 158, 185, 323 Ajugose 391 Acetylcholinesterase 323 Akazie 478, 481 Acetylcoenzym A 44, 376, 378, 442 aktivierte Isopren 44 N-Acetyl-Cystein 265 Alanin 254, 270, 273f, 276, 288, 315 N-Acetyl-D-glucosamin 363 β-Alanin 259 β-Acetyldigoxin 112 Albumin 308 N-Acetylmuraminsäure 363, 365 Albumine 312 O-Acetylsamandarin 234 Alcaligenes faecalis 402 Achras zapota 60 Aldonsäuren 342 Aciclovir 439 Aldosen 335 Aconitum 1 Aldosteron 76, 85, 87 ACP 575 Algin 402 Acrinomycin 260 Alginate 402, 413 Actinomadura 168 Alitam 307 Actinomycin D 306 Alkaloide 143 Actinoplanes 399 Allelopathica 180 Acyl-Carrierprotein 575 Alliin 261 Adamantoylchlorid 352 Allithiamin 426 ADD 95 Allomethylose 341 Adenin 425, 426, 430, 436, 452 Aloin 386 Adenocalymna alliaceum 478 Amanita muscaria 148 Adenosin 432 Amanita phalloides 329 Adenosintriphosphat 442 Amanita strobiliformis 261 Adhatoda vasica 192 Amanitin 329 Adonis vernalis 100, 343 Amantia mappa 148 Adonitol 343 Amaranthenol 41 Adrenalin 145, 146, 147 Amaranthöl 34, 41 Adriamycin 293, 604 AMD 40 676 Sachverzeichnis Amentoflavon 479 anomeren Effekt 339 -
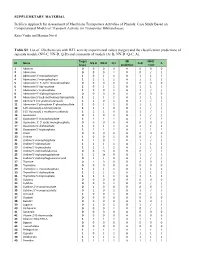
SUPPLEMETARY MATERIAL in Silico Approach for Assessment Of
SUPPLEMETARY MATERIAL In Silico Approach for Assessment of Membrane Transporters Activities of Phenols: Case Study Based on Computational Models of Transport Activity for Transporter Bilitranslocase Katja Venko and Marjana Novič Table S1: List of 120 chemicals with BTL activity experimental values (target) and the classification predictions of separate models (NN-C, NN-D, Q-D) and consensus of models (A+B, NN-D+Q-C, A). Target AB NN-D ID Name NN-D NN-D Q-C A+B A (exp.) prediction + Q-D 1 Adenine 0 0 0 0 A 0 0 0 2 Adenosine 0 0 0 1 B 0 / / 3 Adenosine 3’-monophoshate 1 0 1 1 B 1 1 / 4 Adenosine 5’-monophoshate 1 1 1 1 A 1 1 1 5 Adenosine 3′, 5′-cyclic monophosphate 0 0 0 0 A 0 0 0 6 Adenosine 5′-diphosphate 1 0 1 1 B 1 1 / 7 Adenosine 5′-triphosphate 1 1 0 1 B 1 / / 8 Adenosine-5′-diphosphoglucose 0 0 0 0 A 0 0 0 9 Adenosine 5'-(α,β-methylene) diphosphate 1 1 1 1 A 1 1 1 10 Adenine 9-β-D-arabinofuranoside 1 1 0 1 B 1 / / 11 Adenosine 3′-phosphate 5′-phosphosulfate 1 0 1 1 B 1 1 / 12 S-(5′-Adenosyl)-L-homocysteine 1 1 1 0 B 1 / / 13 S-(5′-Adenosyl)-L-methionine chloride 1 1 1 0 B 1 / / 14 Guanosine 0 1 0 1 B 1 / / 15 Guanosine 5′-monophosphate 1 1 1 1 A 1 1 1 16 Guanosine 3′. -

Buchalter, Leonard, 1922
A PHYTOCHEMICAL INVESTIGATION OF THE ROOTS AND TUBERS OF RUMEX HYMENOSEPALUS FAMILY POLYGONACEAE Item Type text; Dissertation-Reproduction (electronic) Authors Buchalter, Leonard, 1922- Publisher The University of Arizona. Rights Copyright © is held by the author. Digital access to this material is made possible by the University Libraries, University of Arizona. Further transmission, reproduction or presentation (such as public display or performance) of protected items is prohibited except with permission of the author. Download date 29/09/2021 09:03:58 Link to Item http://hdl.handle.net/10150/284768 This dissertation has been microfilmed exactly as received 67-381 BUCHALTER, Leonard, 1922- A PHYTOCHEMICAL INVESTIGATION OF THE ROOTS AND TUBERS OF RUMEX HYMENOSEPALUS FAMILY POLYGONACEAE. University of Arizona, Ph.D., 1966 Chemistry, pharmaceutical University Microfilms, Inc., Ann Arbor, Michigan A PHYTOCHEMICAL INVESTIGATION OF THE ROOTS AND TUBERS OF RUMEX HYMENOSEPALUS FAMILY POLYGONACEAE by Leonard Buchalter A Dissertation Submitted to the Faculty of the COLLEGE OF PHARMACY In Partial Fulfillment of the Requirements For the Degree of DOCTOR OF PHILOSOPHY In The Graduate College THE UNIVERSITY OF ARIZONA 1966 THE UNIVERSITY OF ARIZONA GRADUATE COLLEGE I hereby recommend that this dissertation prepared under my direction by T.pnnar^ Bnp.Vialt.pr entitled A Phyhnr.hprr.i p.al Tnvpsti gflH nn nf thf. Rnohs and Tn-h^-rs nf Piimp-ir HvwiftrinsftT)ali]Br Family Pol vgonaceae, be accepted as fulfilling the dissertation requirement of the degree of nnr.t.rvr of Philosophy ertatioji Director Date After inspection of the dissertation, the following members of the Final Examination Committee concur in its approval and recommend its acceptance:* 7-/3-U / /f *This approval and acceptance is contingent on the candidate's adequate performance and defense of this dissertation at the final oral examination. -

Valeria M. Csizmadia - Budai
PHENOLIC CONSTITUENTS of WESTERN HEMLOCK WOOD (Tsuga Heterophylla (Raf.) Sarg.) by VALERIA M. CSIZMADIA - BUDAI Dipl. Chem. Eng. Polytechnical University of Budapest, 1956 A Thesis Submitted in Partial Fulfilment of the Requirements for the Degree of MASTER OF SCIENCE in the Department of CHEMISTRY We accept this thesis as conforming to the required standard THE UNIVERSITY OF BRITISH COLUMBIA December 1961 In presenting this thesis in partial fulfilment of the requirements for an advanced degree at the University of British Columbia, I agree that the Library shall make it freely available for reference and study. I further agree that permission for extensive copying of this thesis for scholarly purposes may be granted by the Head of my Department or by his representatives. It is understood that copying or publication of this thesis for financial gain shall not be allowed without my written permission. Department of C\-«^^J^*r-^^ The University of British Columbia, Vancouver 8, Canada. Date 2-l> ^^-^^o-^r—y^ ^ nk'iL, (ii) . ABSTRACT The phenolic extractives from western hemlock wood (Tsuga heterophylla (Raf.) Sar.) have been examined The total extractive content of the wood amounted to 1.5$ of the dry weight. A leucoanthocyanidin and two lignans, conidendrin and hydroxymatairesinol, were isolated from the phenolic fraction by precipitation of a methanol solution into peroxide-free ether followed by separation on silicic acid-calcium sulphate chromatobars. The pigment produced by acid treatment of the isolated leucoantho• cyanidin was shown by spectral studies and alkaline degradation to be a mixture of cyanidin and an unidentified anthocyanidin. The two anthocyanidins had identical R^ values in different solvents and similar ultra-violet spectra in ethanol-hydrochloric acid solution but the shift of the absorption maxima caused by addition of aluminium chloride was negligible in the case of the unknown compound and amounted to 30 mji for cyanidin. -

Condensed Tannin Structural Chemistry
Condensed Tannin Structural Chemistry Ann E. Hagerman © March 28, 2002 Proanthocyanidins (condensed tannins) are polymeric flavanoids. The flavanoids are a diverse group of metabolites based on a heterocyclic ring system derived from phenylalanine (B) and polyketide biosynthesis (A). Although the biosynthetic pathways for flavanoid synthesis are 8 B O 7 2 A 6 3 5 4 well understood, the steps leading to condensation and polymerization have not been elucidated. The flavanoid skeleton, the standard letters to identify the rings, and the numbering system are shown here. The most widely studied condensed tannins are based on the flavan-3-ols (-)-epicatechin and (+)- catechin. OH OH OH OH OH O OH O OH OH OH OH epicatechin catechin Flavan-3-ols Addition of a third phenolic group on the B ring yields epigallocatechin and gallocatechin. Much less common are flavan-3-ols with only a single phenolic group on the B ring, para to C-2 (epiafzelechin, afzelechin with stereochemistry corresponding to epicatechin, catechin respectively). Copyright © 2002 by Ann E. Hagerman. All rights reserved. 1 The best characterized condensed tannins are linked via a carbon-carbon bond between C8 of the terminal unit and C4 of the extender. The four common modes of coupling are illustrated by the dimers isolated by Haslam, and originally named B-1, B-2, B-3 and B-4. The more complete names specify the position and stereochemistry of the interflavan bond completely. In addition to these dimers, related dimers linked by C6 of the terminal unit and C4 of the extender have been isolated. OH OH HO O HO O OH OH OH OH OH OH OH OH HO O HO O OH OH OH OH OH OH B-1 B-2 epicatechin-(4β->8)-catechin epicatechin-(4β->8)-epicatechin OH OH HO O HO O OH OH OH OH OH OH OH OH HO O HO O OH OH OH OH OH OH B-3 B-4 catechin-(4α->8)-catechin catechin-(4α->8)-epicatechin Copyright © 2002 by Ann E. -

Genes Controlling Anthocyanidin in the Conversion from Flavanonol to Sepals of Genus Aquilegia
J. Japan. Soc. 1-Iort. Sci. 57(3) : •162-466. 1988. Two Genes Controlling the Conversion from Flavanonol to Anthocyanidin in Sepals of Genus Aquilegia Masao BESS110 Faculty of Agrictcltare, Ta»tagazca t',riversity, Mac/tiida, Tokyo 194 Summary In order to make clear the conversion process from flavanonol to aiithocyanidin, flavonoid components in sepals of .lguilegia flabellata with white flowers (Fw) and .1. hybrida cv. McKana's Giant with creamy white flowers (Mw) were analysed. Isovitextn and populin were identified in both Fw and Mw while leucopelargonidin existed only in Mw. F1 hybrids between Fw and \,Iw showed blue-violet sepals and F, progeny showed two-gene segregation of sepal coloration. Thus the conversion process is controlled by two genes. One recessive gene of Fw controls the reduction of flavanonol and the other recessive gene of Mw controls the dehydration of lcuco- anthocyanidin. Complementation of these two genes enables the synthesis of anthocyani- din in F1 hybrids. anthocyanidin was synthesized. Introduction Materials and Methods Genes controlling flower color expression and the enzymes related to flavonoid metabolism Two strains of /iquilegia flabellata, one of have been studied by many researchers and which had white flowers (Fw) and the other the pathway of anthocyanidin synthesis have had blue-violet ordinal flowers (Fb), and a been well explained by Grisebach(3) and Wong strain with creamy white flowers (Mw) of .l. (11). However, the pathway from flavanonol hybricla cv. McKana's Giant were used for to anthocyanidin is still obscure, although the experiments. Ft and F2 progeny between there have been a few reports on the conver- Fw and Mw were also used for investigation sion process(4, 6, 9, 10).