Protamine Characterization by Top-Down Proteomics: Boosting Proteoform Identification with DBSCAN
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Revista Brasileira De Anestesiologia
Rev BrasBras Anestesiol. Anestesiol. 2013;63(1):163–164 2013;63(1):163-166 REVISTA BRASILEIRA DE ANESTESIOLOGIA Offi cial Publication of the Brazilian Society of Anesthesiology www.sba.com.br/rba/index.asp LETTER TO THE EDITOR Precipitation in Gallipoli: Sugammadex / Amiodarone & Sugammadex / Dobutamine & Sugammadex / Protamine Dear Editor, furosemide (10 mg.mL-1), gentamicin (40 mg.mL-1), glyceryl Sugammadex is a modifi ed gamma cyclodextrin 1-3. Cyclodextrins trinitrate (5 mg.mL-1), heparin (1,000 IU.mL-1), hydrocor- are water soluble cyclic oligosaccharides with a lipophilic tisone (250 mg.mL-1), crystallized insulin (100 IU.mL-1), core. Sugammadex has quickly found a place in clinical use Calcium (Calcium Gluconate Monohydrate 225 mg.10 mL-1 for selective antagonism of neuromuscular blockade with ro- + Calcium levulinate dihitrate 572 mg.10 mL-1), ketamine curonium 1-3. Sugammadex quickly encapsulates steroidal neu- (50 mg.mL-1), levobupivacaine (7.5 mg.mL-1), magnesium romuscular blockers, increasing the amount of encapsulated sulphate (1.2 mEq.mL-1), metamizol sodium (0.5 g.mL-1), steroidal neuromuscular blockers in plasma and separating the methylergobasin maleate (0.2 mg.mL-1), metoclopramide blockers from the nicotinic acetylcholine receptors 1-3. (5 mg.mL-1), metoprolol (1 mg.mL-1), morphine (0.01 g.mL-1), Apart from its use with steroidal neuromuscular block- midazolam (5 mg.mL-1), n-acetylcysteine (100 mg.mL-1), ers, it is known that sugammadex interacts with over 40 naloxone (0.4 mg.mL-1), neostigmine (0.5 mg.mL-1), nitroprus- lipophilic, steroidal and non-steroidal drugs. -
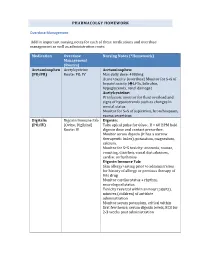
PHARMACOLGY HOMEWORK Overdose Management Add In
PHARMACOLGY HOMEWORK Overdose Management Add in important nursing notes for each of these medications and overdose management as well as administration route. Medication Overdose Nursing Notes (*Homework) Management (Routes) Acetaminophen Acetylcysteine Acetaminophen: (PO/PR) Route: PO, IV Max daily dose: 4000mg Acute toxicity (overdose) Monitor for S+S of hepatotoxicity (éLFTs, bilirubin, hypoglycemia, renal damage) Acetylcysteine: IV infusion: monitor for fluid overload and signs of hyponatremia such as changes in mental status Monitor for S+S of aspiration, bronchospasm, excess secretions Digitalis Digoxin Immune Fab Digoxin: (PO/IV) (Ovine, Digibind) Take apical pulse for 60sec. If < 60 BPM hold Route: IV digoxin dose and contact prescriber. Monitor serum digoxin (it has a narrow therapeutic index), potassium, magnesium, calcium. Monitor for S+S toxicity: anorexia, nausea, vomiting, diarrhea, visual disturbances, cardiac arrhythmias Digoxin Immune Fab: Skin allergy testing prior to administration for history of allergy or previous therapy of this drug Monitor cardiac status + rhythm, neurological status Toxicity reversal within an hour (adults), minutes (children) of antidote administration Monitor serum potassium, critical within first few hours; serum digoxin levels, ECG for 2-3 weeks post administration Heparin Protamine sulfate Heparin: (IV/SC) (IV) Monitor for spontaneous bleeding, thrombocytopenia Monitor aPTT levels Protamine Sulfate: Sudden drop in BP Monitor BP+ P q15-30 min Monitor aPTT Opioid Naloxone (IV-adults, Opioids: -

Estonian Statistics on Medicines 2016 1/41
Estonian Statistics on Medicines 2016 ATC code ATC group / Active substance (rout of admin.) Quantity sold Unit DDD Unit DDD/1000/ day A ALIMENTARY TRACT AND METABOLISM 167,8985 A01 STOMATOLOGICAL PREPARATIONS 0,0738 A01A STOMATOLOGICAL PREPARATIONS 0,0738 A01AB Antiinfectives and antiseptics for local oral treatment 0,0738 A01AB09 Miconazole (O) 7088 g 0,2 g 0,0738 A01AB12 Hexetidine (O) 1951200 ml A01AB81 Neomycin+ Benzocaine (dental) 30200 pieces A01AB82 Demeclocycline+ Triamcinolone (dental) 680 g A01AC Corticosteroids for local oral treatment A01AC81 Dexamethasone+ Thymol (dental) 3094 ml A01AD Other agents for local oral treatment A01AD80 Lidocaine+ Cetylpyridinium chloride (gingival) 227150 g A01AD81 Lidocaine+ Cetrimide (O) 30900 g A01AD82 Choline salicylate (O) 864720 pieces A01AD83 Lidocaine+ Chamomille extract (O) 370080 g A01AD90 Lidocaine+ Paraformaldehyde (dental) 405 g A02 DRUGS FOR ACID RELATED DISORDERS 47,1312 A02A ANTACIDS 1,0133 Combinations and complexes of aluminium, calcium and A02AD 1,0133 magnesium compounds A02AD81 Aluminium hydroxide+ Magnesium hydroxide (O) 811120 pieces 10 pieces 0,1689 A02AD81 Aluminium hydroxide+ Magnesium hydroxide (O) 3101974 ml 50 ml 0,1292 A02AD83 Calcium carbonate+ Magnesium carbonate (O) 3434232 pieces 10 pieces 0,7152 DRUGS FOR PEPTIC ULCER AND GASTRO- A02B 46,1179 OESOPHAGEAL REFLUX DISEASE (GORD) A02BA H2-receptor antagonists 2,3855 A02BA02 Ranitidine (O) 340327,5 g 0,3 g 2,3624 A02BA02 Ranitidine (P) 3318,25 g 0,3 g 0,0230 A02BC Proton pump inhibitors 43,7324 A02BC01 Omeprazole -

Antidote List
Antidote List Recommended minimum amounts for hospital pharmacies to stock Call the Maryland Poison Center for expert advice on the treatment of all poisonings and overdoses: 1-800-222-1222 Poisoning Minimum Stocking Antidote Indication Recommendations Oral: 120 grams Acetylcysteine Acetaminophen IV: 96 grams Crotaline snake Antivenin, snake (CroFab®) 12 vials envenomation 1 vial Black widow spider (Note: Merck limits distribution Antivenin, black widow spider envenomation to cases of confirmed bites with symptoms only) Organophosphate & Atropine carbamate insecticides, nerve 165 mg gases Calcium chloride Calcium channel blockers 10 grams 2.25 grams Calcium disodium EDTA Lead (Available direct from ASD Specialty Healthcare) Hydrofluoric acid 1 kg (powder) Calcium gluconate Calcium channel blockers IV: 30 grams 1 kit Cyanide Antidote Kit Cyanide (or Hydroxocobalamin, see below) Cyproheptadine (Periactin®) Serotonin syndrome 80 mg Neuroleptic malignant Dantrolene syndrome, stimulant-induced 720 mg hyperthermia Deferoxamine mesylate Iron 8 grams (Desferal®) Digoxin Immune FAB Digoxin 20 vials (Digibind®, DigiFab®) Dimercaprol (BAL) Heavy metals 1.5 grams 1 | P a g e Maryland Poison Center Antidote List – continued Poisoning Minimum Stocking Antidote Indication Recommendations DMSA (Succimer, Chemet®) Heavy metals 2000 mg Folic acid Methanol IV: 150 mg Flumazenil (Romazicon®) Benzodiazepines 10 mg Fomepizole (Antizol®) Ethylene glycol, methanol 12 grams Beta blockers, Glucagon 50 mg calcium channel blockers Hydroxocobalamin (Cyanokit®) Cyanide -

Protamine Sulfate Enhances Lipid-Mediated Gene Transfer
Gene Therapy (1997) 4, 961–968 1997 Stockton Press All rights reserved 0969-7128/97 $12.00 Protamine sulfate enhances lipid-mediated gene transfer FL Sorgi1,2, S Bhattacharya1 and L Huang1 1The Laboratory of Drug Targeting, Department of Pharmacology, The University of Pittsburgh School of Medicine, Pittsburgh, PA, USA A polycationic peptide, protamine sulfate, USP, has been USP as a condensation agent was found to be superior to shown to be able to condense plasmid DNA efficiently for poly-L-lysine as well as to various other types of protamine. delivery into several different types of cells in vitro by sev- These differences among various salt forms of protamine eral different types of cationic liposomes. The monovalent appear to be attributable to structural differences between cationic liposomal formulations (DC-Chol and lipofectin) the protamines and not due to differences in the net charge exhibited increased transfection activities comparable to of the molecule. The appearance of lysine residues within that seen with the multivalent cationic liposome formu- the protamine molecule correlate with a reduction in bind- lation, lipofectamine. This suggests that lipofectamine’s ing affinity to plasmid DNA, as well as an observed loss in superior in vitro activity arises from its ability to condense transfection-enhancing activity. This finding sheds light on DNA efficiently and that protamine’s primary role is that of the structural requirements of condensation agents for use a condensation agent, although it also possesses several in gene transfer protocols. Furthermore, protamine sulfate, amino acid sequences resembling that of a nuclear localiz- USP is an FDA-approved compound with a documented ation signal. -

Use of Protamine in Nanopharmaceuticals—A Review
nanomaterials Review Use of Protamine in Nanopharmaceuticals—A Review Ivana Ruseska †, Katja Fresacher †, Christina Petschacher and Andreas Zimmer * Department of Pharmaceutical Technology and Biopharmacy, Institute of Pharmaceutical Sciences, Karl-Franzens-University Graz, Universitätsplatz 1, 8010 Graz, Austria; [email protected] (I.R.); [email protected] (K.F.); [email protected] (C.P.) * Correspondence: [email protected] † Both authors are contributed equally. Abstract: Macromolecular biomolecules are currently dethroning classical small molecule therapeu- tics because of their improved targeting and delivery properties. Protamine-a small polycationic peptide-represents a promising candidate. In nature, it binds and protects DNA against degradation during spermatogenesis due to electrostatic interactions between the negatively charged DNA- phosphate backbone and the positively charged protamine. Researchers are mimicking this technique to develop innovative nanopharmaceutical drug delivery systems, incorporating protamine as a carrier for biologically active components such as DNA or RNA. The first part of this review high- lights ongoing investigations in the field of protamine-associated nanotechnology, discussing the self-assembling manufacturing process and nanoparticle engineering. Immune-modulating prop- erties of protamine are those that lead to the second key part, which is protamine in novel vaccine technologies. Protamine-based RNA delivery systems in vaccines (some belong to the new class of mRNA-vaccines) against infectious disease and their use in cancer treatment are reviewed, and we provide an update on the current state of latest developments with protamine as pharmaceutical Citation: Ruseska, I.; Fresacher, K.; excipient for vaccines. Petschacher, C.; Zimmer, A. Use of Protamine in Nanopharmaceuticals— Keywords: protamine; proticles; nanoparticles; novel vaccine technologies A Review. -

Essential Medications Review
Essential medications for high-quality patient care Quarter 3 2021 update Essential medications – September 2021 As part of the mission to end drug shortages, the Vizient team of pharmacy experts continues to identify essential medications where, if not available, would prove the greatest threat to hospitals’ ability to provide immediate and high-quality patient care. Medications identified as of greatest importance were selected by the Vizient pharmacy team from a comprehensive clinical review of products contained within the World Health Organization’s (WHO) Essential Medicines list, the Advanced Cardiac Life Support (ACLS) and Pediatric Advanced Life Support (PALS) algorithms, and medications included in Vizient member health systems’ critical drug lists. As of this edition, 14 drugs were added to the list creating a total of 251 line items, representing 237 unique drugs and three categories. This includes: • Acute treatment drugs with no alternatives (77 drugs) – Medicines used in acute and critical circumstances to sustain life and for which there are no current alternatives • Chronic treatment drugs with no alternatives (37 drugs) – Products used in chronic disease states or conditions where no alternatives are available (e.g., chemotherapy medications) • High impact drugs (137 drugs) – Medicines for which alternatives are available but may be less clinically desirable and/or are more operationally difficult to use; also reflects drugs where the absence of one medication can affect therapeutically related drugs. Updated quarterly, -

Poison Control Antidote Poster
Antidote Chart (The suggested minimum stocking level is a combination of factors; anticipation of the highest total dose of a drug generally given during a 24 hour period to a 70 kg adult.) General Decontamination Uses Dose Comments Activated Charcoal (without sorbitol) Most ingestions occurring within one hour Adults: 25–100 grams, Children: 1 g/kg May be given in multiple doses depending on ingestant to enhance elimination Activated Charcoal (with sorbitol) May be used as first AC given to patient if presents within Adults: 25–50 grams Should not be used for multiple dose activated charcoal regimens one hour of ingestion Children: Not generally recommended Whole Bowel Irrigation Drugs not bound by charcoal, sustained Adults: 500–2000 ml/hour Nasogastric tube should be used to maintain amount given (Polyethylene Glycol) release formulations, and body stuffers Children: 25 ml/kg/hour Syrup of Ipecac No longer recommended No longer recommended No longer recommended Poisoning Antidote Quantity to Stock* Comments Acetaminophen N-ACETYLYCYSTEINE 10–20% 600 ml (20% Mucomyst®) Because vomiting of the oral NAC is common, the facility should maintain a repeat dose (Mucomyst®) or 1200 ml (10% Mucomyst®) for each patient for initial dosing if continuation of treatment at facility is not anticipated. ACETADOTE® The IV Acetadote® was just approved in February 2004. Call the MAPCC for dosing, ACETYLCYSTEINE Injection for IV use 4 (30 ml) vials of 20% solution precautions and contraindications. Both the oral and IV forms of acetylcysteine should be administered within 8 hours for maximal protection against hepatic injury. Anticholinergic Poisoning PHYSOSTIGMINE (Antilirium®) 2–4 mg* Not generally recommended for children. -

Food and Drugs Act 2005
LAWS OF TRINIDAD AND TOBAGO MINISTRY OF LEGAL AFFAIRS www.legalaffairs.gov.tt FOOD AND DRUGS ACT CHAPTER 30:01 Act 8 of 1960 Amended by 39 of 1968 156/1972 *31 of 1980 16 of 1986 12 of 1987 6 of 1993 16 of 1998 6 of 2005 *See Note on Validation at page 2 Current Authorised Pages Pages Authorised (inclusive) by L.R.O. 1–2 .. 1/2009 3–20 .. 1/2006 21–245 .. 1/2009 L.R.O. 1/2009 UPDATED TO DECEMBER 31ST 2009 LAWS OF TRINIDAD AND TOBAGO MINISTRY OF LEGAL AFFAIRS www.legalaffairs.gov.tt 2 Chap. 30:01 Food and Drugs Index of Subsidiary Legislation Page Food and Drugs Regulations (GN 130/1964) … … … … 25 Official Method Notification (GN 54/1972) … … … … 124 *Approval of New Drugs Notification … … … … … 129 †Withdrawal of Approval of New Drugs Notification (GN 51/1969) … … 200 Fish and Fishery Products Regulations (LN 220/1998)…………201 †This Notification (i.e. 51/1969) has been amended by LNs 99 and 114/1984 which have been omitted. *Note on Approval of New Drugs Notification The list of new drugs set out in the Schedule to this Notification has been consolidated as at 31st December 1977. This list is so voluminous and changes to it so frequent that, especially in view of its very limited use by the general public, it is not practicable to update it annually. The references to the amendments to this list since 31st December 1977 are contained in the Current Consolidated Index of Acts and Subsidiary Legislation. †Note on Withdrawal of Approval of New Drugs Notification For references to the Withdrawal of Approval of New Drugs Notifications subsequent to the year 1969 — See the current Consolidated Index of Acts and Subsidiary Legislation. -

Estonian Statistics on Medicines 2013 1/44
Estonian Statistics on Medicines 2013 DDD/1000/ ATC code ATC group / INN (rout of admin.) Quantity sold Unit DDD Unit day A ALIMENTARY TRACT AND METABOLISM 146,8152 A01 STOMATOLOGICAL PREPARATIONS 0,0760 A01A STOMATOLOGICAL PREPARATIONS 0,0760 A01AB Antiinfectives and antiseptics for local oral treatment 0,0760 A01AB09 Miconazole(O) 7139,2 g 0,2 g 0,0760 A01AB12 Hexetidine(O) 1541120 ml A01AB81 Neomycin+Benzocaine(C) 23900 pieces A01AC Corticosteroids for local oral treatment A01AC81 Dexamethasone+Thymol(dental) 2639 ml A01AD Other agents for local oral treatment A01AD80 Lidocaine+Cetylpyridinium chloride(gingival) 179340 g A01AD81 Lidocaine+Cetrimide(O) 23565 g A01AD82 Choline salicylate(O) 824240 pieces A01AD83 Lidocaine+Chamomille extract(O) 317140 g A01AD86 Lidocaine+Eugenol(gingival) 1128 g A02 DRUGS FOR ACID RELATED DISORDERS 35,6598 A02A ANTACIDS 0,9596 Combinations and complexes of aluminium, calcium and A02AD 0,9596 magnesium compounds A02AD81 Aluminium hydroxide+Magnesium hydroxide(O) 591680 pieces 10 pieces 0,1261 A02AD81 Aluminium hydroxide+Magnesium hydroxide(O) 1998558 ml 50 ml 0,0852 A02AD82 Aluminium aminoacetate+Magnesium oxide(O) 463540 pieces 10 pieces 0,0988 A02AD83 Calcium carbonate+Magnesium carbonate(O) 3049560 pieces 10 pieces 0,6497 A02AF Antacids with antiflatulents Aluminium hydroxide+Magnesium A02AF80 1000790 ml hydroxide+Simeticone(O) DRUGS FOR PEPTIC ULCER AND GASTRO- A02B 34,7001 OESOPHAGEAL REFLUX DISEASE (GORD) A02BA H2-receptor antagonists 3,5364 A02BA02 Ranitidine(O) 494352,3 g 0,3 g 3,5106 A02BA02 Ranitidine(P) -

The Selection of Essential Drugs
This report contains the collective views of an, international group of experts and does not necessarily represent the decisions or the stated policy of the World Health Organization The selection of essential drugs Second report of the WHO Expert Committee World Health Organization Technical Report Series 641 • World Health Organization Geneva 1979 ISBN 92 4 120641 I © World Health Organization 1979 Publications of the World Health Organization enjoy copyright protection in accordance with the provisions of Protocol 2 of the Universal Copyright Convention. For rights of reproduction or translation of WHO publications, in part or in toto, appli cation should be made to the Office of Publications, World Health Organization, Geneva, Switzerland. The World Health Organization welcomes such applications. The designations employed and the presentation of the material in this publication do not imply the expression of any opinion whatsoever on the part of the Secretariat of the World Health Organization concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers- or boundaries. - The mention of specific companies or of certain manufacturers' products does not imply that they are endorsed or recommended by the World Health Organization in preference to others of a similar nature that are not mentioned. Errors and omissions excepted; the names of proprietar j products are distinguished by initial capital letters. PRINTED IN SWlTZERL-\'-iD 79/4523 - 10000 - La Concorde CONTENfS Page I. Introduction . 7 2. General considerations 8 3. Guidelines for the selection of pharmaceutical forms 8 4. Revised model list of essential drugs . -

Amine of Proteins Via Biomimetic Ortho-Quinone-Mediated Oxidation
ARTICLE https://doi.org/10.1038/s41467-021-22654-7 OPEN Modification of N-terminal α-amine of proteins via biomimetic ortho-quinone-mediated oxidation Siyao Wang1,6, Qingqing Zhou1,6, Xiaoping Chen2,6, Rong-Hua Luo3,4,6, Yunxue Li1, Xinliang Liu1, ✉ ✉ Liu-Meng Yang3,4, Yong-Tang Zheng 3,4,7 & Ping Wang 1,5,7 Naturally abundant quinones are important molecules, which play essential roles in various biological processes due to their reduction potential. In contrast to their universality, the 1234567890():,; investigation of reactions between quinones and proteins remains sparse. Herein, we report the development of a convenient strategy to protein modification via a biomimetic quinone- mediated oxidation at the N-terminus. By exploiting unique reactivity of an ortho-quinone reagent, the α-amine of protein N-terminus is oxidized to generate aldo or keto handle for orthogonal conjugation. The applications have been demonstrated using a range of proteins, including myoglobin, ubiquitin and small ubiquitin-related modifier 2 (SUMO2). The effect of this method is further highlighted via the preparation of a series of 17 macrophage inflam- matory protein 1β (MIP-1β) analogs, followed by preliminary anti-HIV activity and cell viability assays, respectively. This method offers an efficient and complementary approach to existing strategies for N-terminal modification of proteins. 1 Shanghai Key Laboratory for Molecular Engineering of Chiral Drugs, School of Chemistry and Chemical Engineering, Frontiers Science Center for Transformative Molecules, Shanghai Jiao Tong University, Shanghai, China. 2 Key Laboratory of Optoelectronic Devices and Systems of Ministry of Education and Guangdong Province College of Physics and Optoelectronic Engineering, Shenzhen University, Shenzhen, China.